Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002491A_C01 KMC002491A_c01
(528 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
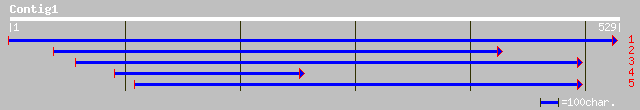
ref|NP_197535.1| ankyrin-repeat protein; protein id: At5g20350.1... 97 9e-20
gb|AAK76360.1|AF272150_1 deliriumA [Dictyostelium discoideum] 46 3e-04
ref|NP_195156.1| putative protein; protein id: At4g34300.1 [Arab... 45 5e-04
sp|P54674|P3K2_DICDI PHOSPHATIDYLINOSITOL 3-KINASE 2 (PI3-KINASE... 44 0.001
ref|NP_700549.1| asparagine-rich antigen [Plasmodium falciparum ... 44 0.002
>ref|NP_197535.1| ankyrin-repeat protein; protein id: At5g20350.1 [Arabidopsis
thaliana]
Length = 592
Score = 97.4 bits (241), Expect = 9e-20
Identities = 52/110 (47%), Positives = 65/110 (58%), Gaps = 1/110 (0%)
Frame = -3
Query: 526 NCSDFLIHGYNEDVEVVEE-LGQSEEGVGMMHMTRNSNLANGDSHSHSEHATGTGNGHHI 350
NCSDFL+ GYNED+E EE Q EG+ MM M RN NL NG NGH
Sbjct: 495 NCSDFLVKGYNEDIECHEEDATQRPEGISMMQMQRNPNLQNG-------------NGHVA 541
Query: 349 INVNSNNSNSKTHHGHVNGHVHSSNCSHANQGKTRNDSTPVGLGLGLGRN 200
I+VN H + HVHS+NCSH++ K+++D+ P+GLGLGL RN
Sbjct: 542 IDVNPT-------HNSQSAHVHSANCSHSHNSKSKSDNVPLGLGLGLSRN 584
>gb|AAK76360.1|AF272150_1 deliriumA [Dictyostelium discoideum]
Length = 817
Score = 45.8 bits (107), Expect = 3e-04
Identities = 20/91 (21%), Positives = 54/91 (58%), Gaps = 2/91 (2%)
Frame = -3
Query: 493 EDVEVVEELGQSEEGVGMMHMTRNSNLANGDSHSHSEHATGTGNGHHIINVNSNNSNSKT 314
ED+++ ++ + G+ + N+N +N +S+S+S + + + ++ N N+NN+N+
Sbjct: 33 EDIDISDQASYHQPGLDKNNNNNNNNNSNSNSNSNSNSNSNSNSNNNNNNNNNNNNNNSN 92
Query: 313 HHGHVNGHVHS--SNCSHANQGKTRNDSTPV 227
++ + N + +S +N +++N + N++ P+
Sbjct: 93 NNNNNNSNNNSNNNNINNSNNNNSNNNNHPI 123
Score = 32.0 bits (71), Expect = 4.6
Identities = 22/83 (26%), Positives = 38/83 (45%), Gaps = 20/83 (24%)
Frame = -3
Query: 424 NSNLANGD---------SHSHSEHATGTGNGHHIINVNSNNSNSKTH----------HGH 302
NSN N + +++++ ++ N ++I N N+NNSN+ H H H
Sbjct: 75 NSNNNNNNNNNNNNNNSNNNNNNNSNNNSNNNNINNSNNNNSNNNNHPINHMQQHHIHQH 134
Query: 301 V-NGHVHSSNCSHANQGKTRNDS 236
+ N H SSN + N N++
Sbjct: 135 LQNYHFQSSNNNSNNNNNNNNNN 157
>ref|NP_195156.1| putative protein; protein id: At4g34300.1 [Arabidopsis thaliana]
gi|7485325|pir||T04776 hypothetical protein F10M10.70 -
Arabidopsis thaliana gi|4455175|emb|CAB36707.1| putative
protein [Arabidopsis thaliana]
gi|7270380|emb|CAB80147.1| putative protein [Arabidopsis
thaliana]
Length = 313
Score = 45.1 bits (105), Expect = 5e-04
Identities = 36/101 (35%), Positives = 51/101 (49%), Gaps = 7/101 (6%)
Frame = -3
Query: 466 GQSEEGVGMMHMTRNSNLANGDSHSHSEHATGTGNGHHIINVNSNNSNS--KTHHGHVNG 293
G S G G ++N G S S S H++GTG+ H+ + SN+S+ TH+GH +G
Sbjct: 62 GGSSWGWGWSSDGTDTNWGWGSS-SGSNHSSGTGSTHNGHSSGSNHSSGTGSTHNGHSSG 120
Query: 292 HVHSSN--CSHANQGKTRNDSTPVG---LGLGLGRNSRSVA 185
HSS+ +H N N S+ VG G G N S+A
Sbjct: 121 SNHSSSTGSTHNNHSSGSNHSSIVGSTHKNHGSGSNHSSIA 161
Score = 35.8 bits (81), Expect = 0.32
Identities = 25/78 (32%), Positives = 38/78 (48%), Gaps = 4/78 (5%)
Frame = -3
Query: 445 GMMHMTRNSNLANGDSHSHSEHATGTGNGHHIINVNSNNSN--SKTHHGHVNGHVHSS-- 278
G H + + NG S S S H++ TG+ H+ + SN+S+ TH H +G HSS
Sbjct: 103 GSNHSSGTGSTHNGHS-SGSNHSSSTGSTHNNHSSGSNHSSIVGSTHKNHGSGSNHSSIA 161
Query: 277 NCSHANQGKTRNDSTPVG 224
+H N S+ +G
Sbjct: 162 GPTHNGHSSGSNHSSIIG 179
>sp|P54674|P3K2_DICDI PHOSPHATIDYLINOSITOL 3-KINASE 2 (PI3-KINASE) (PTDINS-3-KINASE) (PI3K)
gi|7489855|pir||T18273 1-phosphatidylinositol 3-kinase
(EC 2.7.1.137) 2 - slime mold (Dictyostelium discoideum)
gi|733522|gb|AAA85722.1|
phosphatidylinositol-4,5-diphosphate 3-kinase
Length = 1858
Score = 43.9 bits (102), Expect = 0.001
Identities = 25/74 (33%), Positives = 44/74 (58%)
Frame = -3
Query: 427 RNSNLANGDSHSHSEHATGTGNGHHIINVNSNNSNSKTHHGHVNGHVHSSNCSHANQGKT 248
++SN N DS S++ + N ++ N N+NN+N+ ++G+ NG+ +S+N S++N +
Sbjct: 1003 KDSNKENKDSSSNNNNNNNNNNNNNNNNNNNNNNNNNNNNGNNNGN-NSNNNSNSNISRG 1061
Query: 247 RNDSTPVGLGLGLG 206
DS G G G G
Sbjct: 1062 SIDSEGNGSGSGNG 1075
Score = 32.3 bits (72), Expect = 3.5
Identities = 20/89 (22%), Positives = 46/89 (51%), Gaps = 3/89 (3%)
Frame = -3
Query: 490 DVEVVEELGQSEEG-VGMMHMTRN--SNLANGDSHSHSEHATGTGNGHHIINVNSNNSNS 320
D + L +S G +G + N S+ ++ + + + T T N ++ N N+NN+N+
Sbjct: 140 DSTINTPLNRSRSGSIGSKPICNNLTSSSSSSSTTATTPSPTTTSNNNNNNNNNNNNNNN 199
Query: 319 KTHHGHVNGHVHSSNCSHANQGKTRNDST 233
++ + N + +++N ++ N N++T
Sbjct: 200 NNNNNNNNNNNNNNNNNNNNNNNNNNNTT 228
Score = 32.0 bits (71), Expect = 4.6
Identities = 14/59 (23%), Positives = 32/59 (53%)
Frame = -3
Query: 424 NSNLANGDSHSHSEHATGTGNGHHIINVNSNNSNSKTHHGHVNGHVHSSNCSHANQGKT 248
N+N N ++++++ + NG++ N ++NNSNS G ++ + S + ++ T
Sbjct: 1022 NNNNNNNNNNNNNNNNNNNNNGNNNGNNSNNNSNSNISRGSIDSEGNGSGSGNGSEQPT 1080
Score = 31.6 bits (70), Expect = 6.0
Identities = 15/68 (22%), Positives = 37/68 (54%)
Frame = -3
Query: 436 HMTRNSNLANGDSHSHSEHATGTGNGHHIINVNSNNSNSKTHHGHVNGHVHSSNCSHANQ 257
++T +S+ ++ + + S T N ++ N N+NN+N+ ++ + N + +++N ++ N
Sbjct: 163 NLTSSSSSSSTTATTPSPTTTSNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN 222
Query: 256 GKTRNDST 233
ST
Sbjct: 223 NNNNTTST 230
>ref|NP_700549.1| asparagine-rich antigen [Plasmodium falciparum 3D7]
gi|23494939|gb|AAN35273.1|AE014830_17 asparagine-rich
antigen [Plasmodium falciparum 3D7]
Length = 1597
Score = 43.5 bits (101), Expect = 0.002
Identities = 28/122 (22%), Positives = 56/122 (44%), Gaps = 23/122 (18%)
Frame = -3
Query: 514 FLIHGYNE--DVEVVEELGQSE--EGVGMMHMTRNSNLANGDSHSHSEHATGTG---NGH 356
F +HG + D + +L +SE E MHM N+N N +++ H + N H
Sbjct: 584 FHVHGSRKAVDAAITNDLLRSEMEENFNNMHMNHNNNNNNNNNNQHHHQNNNSNIHHNNH 643
Query: 355 HIINVNSN----------------NSNSKTHHGHVNGHVHSSNCSHANQGKTRNDSTPVG 224
H+ N+N N N N+ ++ ++N +++++N ++ N + N++
Sbjct: 644 HMHNMNLNINPNHLGHNMSHNMNHNYNNNNNNMYINNNINNNNNNNNNNSNSNNNNNNNN 703
Query: 223 LG 218
+G
Sbjct: 704 IG 705
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 459,693,703
Number of Sequences: 1393205
Number of extensions: 10263642
Number of successful extensions: 82366
Number of sequences better than 10.0: 894
Number of HSP's better than 10.0 without gapping: 33247
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 55091
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17308240320
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)