Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002470A_C01 KMC002470A_c01
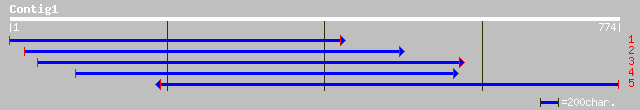
(774 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_201242.1| beta-ureidopropionase; protein id: At5g64370.1 ... 340 1e-92
gb|AAN17428.1| beta-ureidopropionase [Arabidopsis thaliana] gi|2... 340 1e-92
dbj|BAC15525.1| contains ESTs AU094470(E11056),C73573(E11056)~si... 339 3e-92
gb|EAA13946.1| agCP8629 [Anopheles gambiae str. PEST] 296 3e-79
gb|AAK60519.1|AF333186_1 beta-alanine synthase [Dictyostelium di... 294 1e-78
>ref|NP_201242.1| beta-ureidopropionase; protein id: At5g64370.1 [Arabidopsis
thaliana]
Length = 405
Score = 340 bits (873), Expect = 1e-92
Identities = 155/189 (82%), Positives = 171/189 (90%)
Frame = -3
Query: 769 DFNESTYYI*GNTGHPVFETQFGKIGINICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL 590
DFNESTYY+ G+TGHPVFET FGKI +NICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL
Sbjct: 217 DFNESTYYMEGDTGHPVFETVFGKIAVNICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL 276
Query: 589 SEPMWSIEARNGAIANSYFVAAINRVGTETFPNAFTSGDGKPAHADFGHFYGSSYVAAPD 410
SEPMW IEARN AIANSYFV +INRVGTE FPN FTSGDGKP H DFGHFYGSS+ +APD
Sbjct: 277 SEPMWPIEARNAAIANSYFVGSINRVGTEVFPNPFTSGDGKPQHNDFGHFYGSSHFSAPD 336
Query: 409 ASCTPSLSRNRDGLLVTDMDLNLCRQVKDKWGFRMTARYELYEETLANYMKPDFEPQIIG 230
ASCTPSLSR +DGLL++DMDLNLCRQ KDKWGFRMTARYE+Y + LA Y+KPDF+PQ++
Sbjct: 337 ASCTPSLSRYKDGLLISDMDLNLCRQYKDKWGFRMTARYEVYADLLAKYIKPDFKPQVVS 396
Query: 229 DPLLHKKSS 203
DPLLHK S+
Sbjct: 397 DPLLHKNST 405
>gb|AAN17428.1| beta-ureidopropionase [Arabidopsis thaliana]
gi|28193999|gb|AAO33358.1|AF465754_1
N-carbamyl-beta-alanine amidohydrolase [Arabidopsis
thaliana]
Length = 408
Score = 340 bits (873), Expect = 1e-92
Identities = 155/189 (82%), Positives = 171/189 (90%)
Frame = -3
Query: 769 DFNESTYYI*GNTGHPVFETQFGKIGINICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL 590
DFNESTYY+ G+TGHPVFET FGKI +NICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL
Sbjct: 220 DFNESTYYMEGDTGHPVFETVFGKIAVNICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL 279
Query: 589 SEPMWSIEARNGAIANSYFVAAINRVGTETFPNAFTSGDGKPAHADFGHFYGSSYVAAPD 410
SEPMW IEARN AIANSYFV +INRVGTE FPN FTSGDGKP H DFGHFYGSS+ +APD
Sbjct: 280 SEPMWPIEARNAAIANSYFVGSINRVGTEVFPNPFTSGDGKPQHNDFGHFYGSSHFSAPD 339
Query: 409 ASCTPSLSRNRDGLLVTDMDLNLCRQVKDKWGFRMTARYELYEETLANYMKPDFEPQIIG 230
ASCTPSLSR +DGLL++DMDLNLCRQ KDKWGFRMTARYE+Y + LA Y+KPDF+PQ++
Sbjct: 340 ASCTPSLSRYKDGLLISDMDLNLCRQYKDKWGFRMTARYEVYADLLAKYIKPDFKPQVVS 399
Query: 229 DPLLHKKSS 203
DPLLHK S+
Sbjct: 400 DPLLHKNST 408
>dbj|BAC15525.1| contains ESTs AU094470(E11056),C73573(E11056)~similar to
beta-ureidopropionase [Oryza sativa (japonica
cultivar-group)]
Length = 413
Score = 339 bits (869), Expect = 3e-92
Identities = 153/188 (81%), Positives = 171/188 (90%)
Frame = -3
Query: 769 DFNESTYYI*GNTGHPVFETQFGKIGINICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL 590
DFNESTYY+ GNTGHPVFET +GKIG+NICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL
Sbjct: 226 DFNESTYYMEGNTGHPVFETAYGKIGVNICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL 285
Query: 589 SEPMWSIEARNGAIANSYFVAAINRVGTETFPNAFTSGDGKPAHADFGHFYGSSYVAAPD 410
SEPMW IEARN AIANSYFV +INRVGTE FPN FTSGDGKP HADFGHFYGSS+ +APD
Sbjct: 286 SEPMWPIEARNAAIANSYFVGSINRVGTEVFPNPFTSGDGKPQHADFGHFYGSSHFSAPD 345
Query: 409 ASCTPSLSRNRDGLLVTDMDLNLCRQVKDKWGFRMTARYELYEETLANYMKPDFEPQIIG 230
ASCTPSLSR RDGL+++DMDLNLCRQ+KDKWGFRMTARY+ Y L+ Y+KPDF+PQ+I
Sbjct: 346 ASCTPSLSRYRDGLMISDMDLNLCRQIKDKWGFRMTARYDTYASLLSEYLKPDFKPQVIV 405
Query: 229 DPLLHKKS 206
DPL++K +
Sbjct: 406 DPLINKSA 413
>gb|EAA13946.1| agCP8629 [Anopheles gambiae str. PEST]
Length = 392
Score = 296 bits (757), Expect = 3e-79
Identities = 137/179 (76%), Positives = 153/179 (84%)
Frame = -3
Query: 769 DFNESTYYI*GNTGHPVFETQFGKIGINICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL 590
DFNESTYY G+TGHPVF+TQFG+I INICYGRHHP NW+ FG+NGAEIVFNPSATVG L
Sbjct: 211 DFNESTYYFEGDTGHPVFDTQFGRIAINICYGRHHPQNWMMFGVNGAEIVFNPSATVGAL 270
Query: 589 SEPMWSIEARNGAIANSYFVAAINRVGTETFPNAFTSGDGKPAHADFGHFYGSSYVAAPD 410
SEP+W IEARN AIANSYF AINRVGTE FPN FTSG+G PAH DFG FYGSSYVAAPD
Sbjct: 271 SEPLWGIEARNAAIANSYFTVAINRVGTEVFPNEFTSGNGLPAHKDFGPFYGSSYVAAPD 330
Query: 409 ASCTPSLSRNRDGLLVTDMDLNLCRQVKDKWGFRMTARYELYEETLANYMKPDFEPQII 233
S TP LSR++DGLLV +MDLNLCRQ+KD WGF+MT R LY E+LA +KPD++PQII
Sbjct: 331 GSRTPGLSRDKDGLLVVEMDLNLCRQIKDFWGFQMTQRLPLYAESLAKAVKPDYKPQII 389
>gb|AAK60519.1|AF333186_1 beta-alanine synthase [Dictyostelium discoideum]
gi|28829985|gb|AAO52475.1| similar to Dictyostelium
discoideum (Slime mold). Beta-alanine synthase (EC
3.5.1.6)
Length = 391
Score = 294 bits (752), Expect = 1e-78
Identities = 133/182 (73%), Positives = 155/182 (85%)
Frame = -3
Query: 769 DFNESTYYI*GNTGHPVFETQFGKIGINICYGRHHPLNWLAFGLNGAEIVFNPSATVGEL 590
DFNESTYY+ GHPVFET +GKI INICYGRHH LNWLA+GLNGAEIVFNPSATVGEL
Sbjct: 207 DFNESTYYMESTLGHPVFETIYGKIAINICYGRHHNLNWLAYGLNGAEIVFNPSATVGEL 266
Query: 589 SEPMWSIEARNGAIANSYFVAAINRVGTETFPNAFTSGDGKPAHADFGHFYGSSYVAAPD 410
SEPMW +EARN A+ N+YFV +INRVGTE FPN FTSG+GKPAH DFGHFYGSSY ++PD
Sbjct: 267 SEPMWGVEARNAAMTNNYFVGSINRVGTEHFPNEFTSGNGKPAHKDFGHFYGSSYFSSPD 326
Query: 409 ASCTPSLSRNRDGLLVTDMDLNLCRQVKDKWGFRMTARYELYEETLANYMKPDFEPQIIG 230
CTPSLSR DGL ++++DLNLC+QVKDKW F+MTARYELY + L +Y+ P+++P II
Sbjct: 327 NCCTPSLSRVSDGLNISEVDLNLCQQVKDKWNFQMTARYELYAKFLTDYINPNYQPNIIK 386
Query: 229 DP 224
DP
Sbjct: 387 DP 388
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 691,874,425
Number of Sequences: 1393205
Number of extensions: 15801700
Number of successful extensions: 34969
Number of sequences better than 10.0: 219
Number of HSP's better than 10.0 without gapping: 33679
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34894
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38095156112
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)