Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002457A_C01 KMC002457A_c01
(1014 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
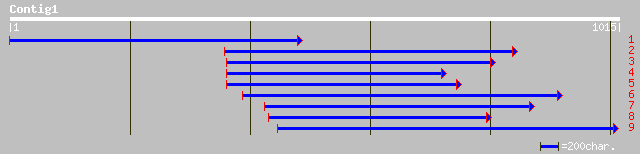
pir||S47081 hypothetical 21.4K protein - chickpea gi|499060|emb|... 74 5e-22
ref|NP_563693.1| Expressed protein; protein id: At1g03900.1, sup... 93 6e-18
gb|AAK13159.1|AC078829_11 unknown protein [Oryza sativa (japonic... 89 1e-16
gb|AAK68815.1| Unknown protein [Arabidopsis thaliana] gi|1837752... 68 3e-10
pir||G86169 hypothetical protein [imported] - Arabidopsis thalia... 65 1e-09
>pir||S47081 hypothetical 21.4K protein - chickpea gi|499060|emb|CAA56128.1|
unnamed protein product [Cicer arietinum]
Length = 182
Score = 73.6 bits (179), Expect(2) = 5e-22
Identities = 40/60 (66%), Positives = 45/60 (74%)
Frame = -3
Query: 982 LLPHRVGEGNSGLLFHPLLMILLLLGLFPPLMDLDLKGHTRV*GILLTLYQIFLNYRRIF 803
LLPH+ + S LLFHP MILLLLGL P +DL LKGHT++ ILL LYQIFLN RRIF
Sbjct: 43 LLPHQAEQEKSDLLFHPHRMILLLLGLLLPCLDLVLKGHTKL-DILLILYQIFLNCRRIF 101
Score = 53.9 bits (128), Expect(2) = 5e-22
Identities = 31/59 (52%), Positives = 39/59 (65%)
Frame = -2
Query: 749 LYIYMSMLTMRLLLLSDFFGSLF*EKKQWRKLKKGGVISKVSARYQGILCLEGSPGNMY 573
L IY S+LTMRLLL +DFFG L K+ + ++ GVIS VS RY+ I LE GN+Y
Sbjct: 118 LIIYTSILTMRLLLFADFFGVLHLRKEIVEETEERGVISIVSKRYRVIRYLEECSGNIY 176
Score = 38.9 bits (89), Expect = 0.13
Identities = 25/50 (50%), Positives = 28/50 (56%)
Frame = -1
Query: 795 SHFRIDHSFRMGSLLIIYIHEHVNYEAFVVV*FFWFFILRKETVEETEER 646
SH I + RMGSLLIIY + + FF LRKE VEETEER
Sbjct: 104 SHSWISNCSRMGSLLIIYT-SILTMRLLLFADFFGVLHLRKEIVEETEER 152
>ref|NP_563693.1| Expressed protein; protein id: At1g03900.1, supported by cDNA:
gi_14596174, supported by cDNA: gi_18377527 [Arabidopsis
thaliana]
Length = 272
Score = 93.2 bits (230), Expect = 6e-18
Identities = 54/91 (59%), Positives = 61/91 (66%), Gaps = 4/91 (4%)
Frame = -2
Query: 1013 AATPKPKVLSLAPPPSGGGKLRSPLPPPPNDPVAARIVSTTHGSGPKGTHEGVRHSTDAL 834
+ T KPK L+LAPPP G RSPLPPPPNDPVA+RI S G E R+ + L
Sbjct: 191 SGTGKPKPLALAPPPKAAGVTRSPLPPPPNDPVASRIAS-------DGCKESRRN--EPL 241
Query: 833 SDLSQLQKNLPSTATSGST----TASGWAAF 753
SDLSQL+KNLPSTA SGS+ ASGWAAF
Sbjct: 242 SDLSQLKKNLPSTAGSGSSKSTGAASGWAAF 272
>gb|AAK13159.1|AC078829_11 unknown protein [Oryza sativa (japonica cultivar-group)]
gi|16905180|gb|AAL31050.1|AC078893_13 unknown protein
[Oryza sativa]
Length = 287
Score = 89.0 bits (219), Expect = 1e-16
Identities = 47/89 (52%), Positives = 62/89 (68%), Gaps = 3/89 (3%)
Frame = -2
Query: 1010 ATPKPKV-LSLAPPPSGGGKLRSPLPPPPNDPVAARIVSTTHGSGPKGTHEGVRHSTDAL 834
A+ KPK + LAPPP GKLRSPLPPPPNDP AAR+ S ++ +G + E + ++DA
Sbjct: 200 ASAKPKASMLLAPPPGSAGKLRSPLPPPPNDPAAARMNSGSN-AGIRAPKEPAKRNSDAF 258
Query: 833 SDLSQLQKNLPSTATSGST--TASGWAAF 753
SDLS +++NLPS+ S T T +GWAAF
Sbjct: 259 SDLSAMKQNLPSSTESAQTKSTGAGWAAF 287
>gb|AAK68815.1| Unknown protein [Arabidopsis thaliana] gi|18377528|gb|AAL66930.1|
unknown protein [Arabidopsis thaliana]
Length = 276
Score = 67.8 bits (164), Expect = 3e-10
Identities = 38/69 (55%), Positives = 45/69 (65%)
Frame = -2
Query: 1013 AATPKPKVLSLAPPPSGGGKLRSPLPPPPNDPVAARIVSTTHGSGPKGTHEGVRHSTDAL 834
+ T KPK L+LAPPP G RSPLPPPPNDPVA+RI S G E R+ + L
Sbjct: 191 SGTGKPKPLALAPPPKAAGVTRSPLPPPPNDPVASRIAS-------DGCKESRRN--EPL 241
Query: 833 SDLSQLQKN 807
SDLSQL+K+
Sbjct: 242 SDLSQLKKS 250
>pir||G86169 hypothetical protein [imported] - Arabidopsis thaliana
gi|4204313|gb|AAD10694.1| Hypothetical protein
[Arabidopsis thaliana]
Length = 580
Score = 65.5 bits (158), Expect = 1e-09
Identities = 37/67 (55%), Positives = 43/67 (63%)
Frame = -2
Query: 1013 AATPKPKVLSLAPPPSGGGKLRSPLPPPPNDPVAARIVSTTHGSGPKGTHEGVRHSTDAL 834
+ T KPK L+LAPPP G RSPLPPPPNDPVA+RI S G E R+ + L
Sbjct: 212 SGTGKPKPLALAPPPKAAGVTRSPLPPPPNDPVASRIAS-------DGCKESRRN--EPL 262
Query: 833 SDLSQLQ 813
SDLSQL+
Sbjct: 263 SDLSQLK 269
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 926,469,846
Number of Sequences: 1393205
Number of extensions: 22878692
Number of successful extensions: 94866
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 76850
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 92414
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 58773479151
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)