Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002453A_C01 KMC002453A_c01
(583 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
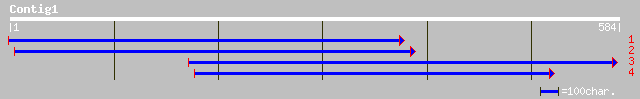
gb|AAK69401.1|AF274564_1 immediate-early fungal elicitor protein... 106 2e-22
ref|NP_566184.1| expressed protein; protein id: At3g02840.1, sup... 95 7e-19
gb|AAM65334.1| unknown [Arabidopsis thaliana] 95 7e-19
dbj|BAB86896.1| syringolide-induced protein 13-1-1 [Glycine max] 89 3e-17
gb|AAN04506.1| Hypothetical protein [Oryza sativa (japonica cult... 82 4e-15
>gb|AAK69401.1|AF274564_1 immediate-early fungal elicitor protein CMPG1 [Petroselinum
crispum] gi|14582202|gb|AAK69402.1|AF274565_1
immediate-early fungal elicitor protein CMPG1
[Petroselinum crispum]
Length = 442
Score = 106 bits (265), Expect = 2e-22
Identities = 58/90 (64%), Positives = 71/90 (78%), Gaps = 1/90 (1%)
Frame = -1
Query: 583 LRVSALSTGYSVSAMWKLCKFGEKKDDDDEGRVLVEALQVGAFQKLLLVLQVGCTEETKE 404
LRVS L+T +SVS +WKL K D ++G V+VEAL+VGAFQKLLL+LQ GC E+ K+
Sbjct: 357 LRVSDLATEFSVSIVWKL----SKNDKSEDGGVIVEALRVGAFQKLLLLLQFGCNEKIKD 412
Query: 403 RATELLKLLNPYRADLECIDS-DFKNLKRS 317
+ATELLKLLN +R LECIDS DFKNLKR+
Sbjct: 413 KATELLKLLNLHRDKLECIDSMDFKNLKRT 442
>ref|NP_566184.1| expressed protein; protein id: At3g02840.1, supported by cDNA:
38495. [Arabidopsis thaliana]
gi|6728978|gb|AAF26976.1|AC018363_21 unknown protein
[Arabidopsis thaliana] gi|26452125|dbj|BAC43151.1|
unknown protein [Arabidopsis thaliana]
gi|27311855|gb|AAO00893.1| expressed protein
[Arabidopsis thaliana]
Length = 379
Score = 94.7 bits (234), Expect = 7e-19
Identities = 54/95 (56%), Positives = 72/95 (74%), Gaps = 5/95 (5%)
Frame = -1
Query: 583 LRVSALSTGYSVSAMWKLCKFGEKKDDDDEGRVLVEALQVGAFQKLLLVLQVGCTEETKE 404
LRVS L+T SVS +WKL K KKD + + R+LVEALQVGAF+KLL++LQVGC ++TKE
Sbjct: 287 LRVSDLATQCSVSILWKLWK--NKKDGECDDRLLVEALQVGAFEKLLVLLQVGCEDKTKE 344
Query: 403 RATELLKLLNPYRADLE---CIDSD--FKNLKRSF 314
+A+ELL+ LN R ++E C+DS KN+K+SF
Sbjct: 345 KASELLRNLNRCRNEIEKTNCVDSSMHLKNVKKSF 379
>gb|AAM65334.1| unknown [Arabidopsis thaliana]
Length = 379
Score = 94.7 bits (234), Expect = 7e-19
Identities = 54/95 (56%), Positives = 72/95 (74%), Gaps = 5/95 (5%)
Frame = -1
Query: 583 LRVSALSTGYSVSAMWKLCKFGEKKDDDDEGRVLVEALQVGAFQKLLLVLQVGCTEETKE 404
LRVS L+T SVS +WKL K KKD + + R+LVEALQVGAF+KLL++LQVGC ++TKE
Sbjct: 287 LRVSDLATQCSVSILWKLWK--NKKDGECDDRLLVEALQVGAFEKLLVLLQVGCEDKTKE 344
Query: 403 RATELLKLLNPYRADLE---CIDSD--FKNLKRSF 314
+A+ELL+ LN R ++E C+DS KN+K+SF
Sbjct: 345 KASELLRNLNRCRNEIEKTNCVDSSMHLKNVKKSF 379
>dbj|BAB86896.1| syringolide-induced protein 13-1-1 [Glycine max]
Length = 431
Score = 89.4 bits (220), Expect = 3e-17
Identities = 51/92 (55%), Positives = 66/92 (71%), Gaps = 2/92 (2%)
Frame = -1
Query: 583 LRVSALSTGYSVSAMWKLCKFGEKKDDDDEGRVLVEALQVGAFQKLLLVLQVGCTEETKE 404
LRVS L++G++VS + K+C D +EG +L+EALQVG FQKL ++LQVGC E TKE
Sbjct: 347 LRVSPLASGFAVSILRKIC------DKREEG-ILIEALQVGLFQKLSVLLQVGCDESTKE 399
Query: 403 RATELLKLLNPYRADLECIDS--DFKNLKRSF 314
AT LLKLLN YR EC DS +F++LK+ F
Sbjct: 400 NATGLLKLLNGYRNKAECTDSSLNFEHLKKPF 431
>gb|AAN04506.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 460
Score = 82.4 bits (202), Expect = 4e-15
Identities = 48/96 (50%), Positives = 64/96 (66%), Gaps = 9/96 (9%)
Frame = -1
Query: 580 RVSALSTGYSVSAMWKLCKFGEKKDDDDEGR------VLVEALQVGAFQKLLLVLQVGCT 419
RVS +T VSA+ ++CK DDD+ G +VEA+QVGAFQK++++LQVGC
Sbjct: 363 RVSDTATELVVSALHRICKKWHDGDDDEVGSPAARRSAVVEAVQVGAFQKVMMLLQVGCR 422
Query: 418 EETKERATELLKLLNPY--RADLECIDS-DFKNLKR 320
+ TKE+ATELLKL+ Y R CID+ DF+ LKR
Sbjct: 423 DATKEKATELLKLMIKYETRGGAHCIDAMDFRGLKR 458
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 443,602,100
Number of Sequences: 1393205
Number of extensions: 8467374
Number of successful extensions: 23584
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 22642
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23515
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)