Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
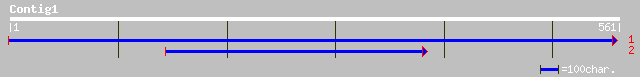
Query= KMC002439A_C01 KMC002439A_c01
(554 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_649074.1| CG14076-PA [Drosophila melanogaster] gi|7293845... 34 1.0
emb|CAA33190.1| MURF2 protein (AA 1-348) [Crithidia fasciculata] 34 1.0
gb|AAA73419.1| cytochrome C oxidase subunit III 33 2.3
gb|EAA01716.1| agCP12327 [Anopheles gambiae str. PEST] 32 5.2
ref|NP_609801.2| CG31738-PB [Drosophila melanogaster] gi|2294664... 32 6.8
>ref|NP_649074.1| CG14076-PA [Drosophila melanogaster] gi|7293845|gb|AAF49210.1|
CG14076-PA [Drosophila melanogaster]
Length = 627
Score = 34.3 bits (77), Expect = 1.0
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 1/59 (1%)
Frame = -1
Query: 374 MVV*LYLGHNCHPLMEFVDCHF-IRNHETWLMWYSYAFDIYFCWFSLKTTLRCLEYVDS 201
M+V Y H H LM V CH I + W +W + + ++ S +++LR L++VD+
Sbjct: 1 MLVAYYRHHGVHSLM-LVVCHTDIADFRLWKLWQHFNLNNFYVQVSTESSLRDLQHVDA 58
>emb|CAA33190.1| MURF2 protein (AA 1-348) [Crithidia fasciculata]
Length = 347
Score = 34.3 bits (77), Expect = 1.0
Identities = 25/80 (31%), Positives = 40/80 (49%)
Frame = -3
Query: 261 YLFLLVLLENYIEMFRICGLFSLHYLFISMRFFLVRF**GISSKCLAFKEYMISCFDVIL 82
++F V+ +I +F + + + F+ + F+V F G+ S L F Y I + IL
Sbjct: 33 FVFDFVVCITFIFVFVLGFFLRIFFSFVFVLLFIVFF--GLFSLALTFTGYYIY-YIYIL 89
Query: 81 FVFSSHLSLCIGLKYLLYFI 22
+ F C GL Y+LYFI
Sbjct: 90 YNFICFF-FCFGLSYILYFI 108
>gb|AAA73419.1| cytochrome C oxidase subunit III
Length = 288
Score = 33.1 bits (74), Expect = 2.3
Identities = 26/98 (26%), Positives = 45/98 (45%), Gaps = 12/98 (12%)
Frame = -3
Query: 273 LCI*YLFLLVLLENYIEMFRICGLFSLHYLFISMRFFLVRF**GISSKCLAFKEYMISCF 94
+C+ Y L++L E ++++FR GL+ L + ++ V F + S+ L F + F
Sbjct: 55 VCLLYCVLIILSEAFVDIFR--GLYDTSTLLRACQYCFVWF---VLSELLLFASFFYVVF 109
Query: 93 DVIL-------FVFSSHLSLC-----IGLKYLLYFISI 16
++L FVF L C G + YF+ I
Sbjct: 110 GLVLFMVCEFAFVFCLPLLFCCLLCDYGFVFYWYFLDI 147
>gb|EAA01716.1| agCP12327 [Anopheles gambiae str. PEST]
Length = 187
Score = 32.0 bits (71), Expect = 5.2
Identities = 14/42 (33%), Positives = 23/42 (54%)
Frame = +2
Query: 254 NKYQMHNYTTSARSRGSL*NGSPQIPSKGGNCGQGTTTQPSA 379
N +N T A++ G +G P +PS G G+ +T +PS+
Sbjct: 32 NSSSANNSTVQAQATGGTGSGEPVLPSTTGTTGRSSTLRPSS 73
>ref|NP_609801.2| CG31738-PB [Drosophila melanogaster] gi|22946643|gb|AAF53546.3|
CG31738-PB [Drosophila melanogaster]
Length = 1700
Score = 31.6 bits (70), Expect = 6.8
Identities = 17/54 (31%), Positives = 28/54 (51%)
Frame = +3
Query: 219 TSQCSFQGEPTKINIKCITIPHQPGLVVPYKMAVHKFHQRVAIVAKVQLHNHRP 380
+S CS P + + P Q +VP A+H Q++A++A +Q H+H P
Sbjct: 60 SSNCSLPASPPLL--QQTATPPQGAQIVPPVCALHHPQQQLALMAAMQHHHHLP 111
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,395,401
Number of Sequences: 1393205
Number of extensions: 11078908
Number of successful extensions: 26137
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 25432
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26133
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)