Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
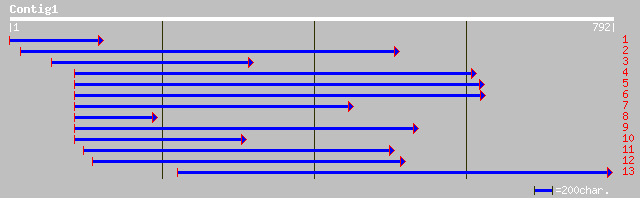
Query= KMC002434A_C01 KMC002434A_c01
(791 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_196828.2| putative protein; protein id: At5g13240.1, supp... 173 3e-42
pir||T48571 hypothetical protein T31B5.60 - Arabidopsis thaliana... 173 3e-42
gb|EAA12877.1| agCP11035 [Anopheles gambiae str. PEST] 75 1e-12
ref|NP_115648.1| homolog of yeast MAF1; hypothetical protein DKF... 73 4e-12
gb|AAH46951.1| Similar to RIKEN cDNA 1110068E11 gene [Xenopus la... 73 5e-12
>ref|NP_196828.2| putative protein; protein id: At5g13240.1, supported by cDNA:
gi_19347945 [Arabidopsis thaliana]
gi|19347946|gb|AAL86308.1| unknown protein [Arabidopsis
thaliana] gi|21689797|gb|AAM67542.1| unknown protein
[Arabidopsis thaliana]
Length = 224
Score = 173 bits (438), Expect = 3e-42
Identities = 82/124 (66%), Positives = 98/124 (78%)
Frame = -3
Query: 789 AHQFFTEESWDSFKQVFDSYMFEASKEWDETVEGLSLLDTLLKAIDEVVKLADCEIYGYV 610
AHQFF+EESWD+FKQ+F++YMFEASKEW E E SLL+ + KA+DEVVKLA+CEIY Y
Sbjct: 101 AHQFFSEESWDTFKQIFNNYMFEASKEWTERNEDGSLLEVIYKALDEVVKLAECEIYVYN 160
Query: 609 PDYEADPLLDSGAMWSFNFLFYNRKLKRVVTFHFSCFSNLITDGFLADEFHHDYDEQIFA 430
P+ ADP L+ GA+WSF FLFYNRKLKRV F FSC SNL D FL D ++ DE+IFA
Sbjct: 161 PNPNADPFLEEGAIWSFCFLFYNRKLKRVAGFRFSCTSNLANDAFLTDSPPYEEDEEIFA 220
Query: 429 DMDI 418
DMD+
Sbjct: 221 DMDM 224
>pir||T48571 hypothetical protein T31B5.60 - Arabidopsis thaliana
gi|7529279|emb|CAB86631.1| putative protein [Arabidopsis
thaliana]
Length = 283
Score = 173 bits (438), Expect = 3e-42
Identities = 82/124 (66%), Positives = 98/124 (78%)
Frame = -3
Query: 789 AHQFFTEESWDSFKQVFDSYMFEASKEWDETVEGLSLLDTLLKAIDEVVKLADCEIYGYV 610
AHQFF+EESWD+FKQ+F++YMFEASKEW E E SLL+ + KA+DEVVKLA+CEIY Y
Sbjct: 160 AHQFFSEESWDTFKQIFNNYMFEASKEWTERNEDGSLLEVIYKALDEVVKLAECEIYVYN 219
Query: 609 PDYEADPLLDSGAMWSFNFLFYNRKLKRVVTFHFSCFSNLITDGFLADEFHHDYDEQIFA 430
P+ ADP L+ GA+WSF FLFYNRKLKRV F FSC SNL D FL D ++ DE+IFA
Sbjct: 220 PNPNADPFLEEGAIWSFCFLFYNRKLKRVAGFRFSCTSNLANDAFLTDSPPYEEDEEIFA 279
Query: 429 DMDI 418
DMD+
Sbjct: 280 DMDM 283
>gb|EAA12877.1| agCP11035 [Anopheles gambiae str. PEST]
Length = 288
Score = 74.7 bits (182), Expect = 1e-12
Identities = 31/76 (40%), Positives = 47/76 (61%)
Frame = -3
Query: 669 LLKAIDEVVKLADCEIYGYVPDYEADPLLDSGAMWSFNFLFYNRKLKRVVTFHFSCFSNL 490
L I++ + L DC+IY Y PD +DP + G +WSFN+ FYN+KLKR+V FH +++
Sbjct: 211 LWTTIEDEISLKDCDIYSYNPDLNSDPFGEPGCLWSFNYFFYNKKLKRIVFFHCRAINSV 270
Query: 489 ITDGFLADEFHHDYDE 442
D + +F + DE
Sbjct: 271 YVDSGFSSDFAMEDDE 286
>ref|NP_115648.1| homolog of yeast MAF1; hypothetical protein DKFZp586G1123 [Homo
sapiens] gi|12053369|emb|CAB66871.1| hypothetical
protein [Homo sapiens]
gi|15559429|gb|AAH14082.1|AAH14082 homolog of yeast MAF1
[Homo sapiens] gi|17511721|gb|AAH18714.1|AAH18714
homolog of yeast MAF1 [Homo sapiens]
gi|21411503|gb|AAH31273.1| homolog of yeast MAF1 [Homo
sapiens]
Length = 256
Score = 73.2 bits (178), Expect = 4e-12
Identities = 38/98 (38%), Positives = 57/98 (57%)
Frame = -3
Query: 789 AHQFFTEESWDSFKQVFDSYMFEASKEWDETVEGLSLLDTLLKAIDEVVKLADCEIYGYV 610
+H+F E S + +F A +E + L L A+DE + LA+C+IY Y
Sbjct: 115 SHEFSREPSLSWVVNAVNCSLFSAVRE-----DFKDLKPQLWNAVDEEICLAECDIYSYN 169
Query: 609 PDYEADPLLDSGAMWSFNFLFYNRKLKRVVTFHFSCFS 496
PD ++DP + G++WSFN+ FYN++LKR+V FSC S
Sbjct: 170 PDLDSDPFGEDGSLWSFNYFFYNKRLKRIV--FFSCRS 205
>gb|AAH46951.1| Similar to RIKEN cDNA 1110068E11 gene [Xenopus laevis]
Length = 245
Score = 72.8 bits (177), Expect = 5e-12
Identities = 39/98 (39%), Positives = 56/98 (56%)
Frame = -3
Query: 789 AHQFFTEESWDSFKQVFDSYMFEASKEWDETVEGLSLLDTLLKAIDEVVKLADCEIYGYV 610
AH+F E S + +S + A + + SL L A+DE + L++C+IY Y
Sbjct: 114 AHEFSREPSLNWVVNAVNSSLVSALGD-----DFTSLRPNLWDAVDEEINLSECDIYSYN 168
Query: 609 PDYEADPLLDSGAMWSFNFLFYNRKLKRVVTFHFSCFS 496
PD + DP + G +WSFN+ FYN+KLKR+V FSC S
Sbjct: 169 PDLDCDPFGEDGNLWSFNYFFYNKKLKRIV--FFSCRS 204
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 646,162,573
Number of Sequences: 1393205
Number of extensions: 13585199
Number of successful extensions: 33341
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 31859
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33289
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 39775824764
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)