Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002429A_C01 KMC002429A_c01
(590 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
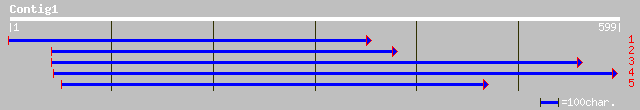
ref|NP_568577.1| DegP protease; protein id: At5g40200.1, support... 108 4e-23
gb|AAB60923.1| F5I14.16 [Arabidopsis thaliana] 50 2e-05
ref|NP_564856.1| DegP protease; protein id: At1g65630.1 [Arabido... 50 2e-05
ref|NP_566552.1| DegP protease; protein id: At3g16550.1 [Arabido... 50 2e-05
dbj|BAB01154.1| contains similarity to serine protease~gene_id:M... 50 2e-05
>ref|NP_568577.1| DegP protease; protein id: At5g40200.1, supported by cDNA:
gi_15028146 [Arabidopsis thaliana]
gi|10177507|dbj|BAB10901.1| serine protease-like protein
[Arabidopsis thaliana] gi|15028147|gb|AAK76697.1|
putative DegP protease [Arabidopsis thaliana]
gi|23296815|gb|AAN13177.1| putative DegP protease
[Arabidopsis thaliana]
Length = 592
Score = 108 bits (271), Expect = 4e-23
Identities = 52/62 (83%), Positives = 56/62 (89%)
Frame = -3
Query: 588 GKPVKNLKSLAAMVESCRDEYLKFNLEYDQIVVLRTKTAKAATLDILATHCIPSAMSDDL 409
GKPVKNLK LA MVE+C DEY+KFNL+YDQIVVL TKTAK ATLDIL THCIPSAMSDDL
Sbjct: 527 GKPVKNLKGLAGMVENCEDEYMKFNLDYDQIVVLDTKTAKEATLDILTTHCIPSAMSDDL 586
Query: 408 KS 403
K+
Sbjct: 587 KT 588
>gb|AAB60923.1| F5I14.16 [Arabidopsis thaliana]
Length = 487
Score = 50.4 bits (119), Expect = 2e-05
Identities = 27/61 (44%), Positives = 39/61 (63%)
Frame = -3
Query: 588 GKPVKNLKSLAAMVESCRDEYLKFNLEYDQIVVLRTKTAKAATLDILATHCIPSAMSDDL 409
G V NLK L +VE C E ++ +LE D+++ L K+AK T IL + IPSA+S+DL
Sbjct: 410 GVQVHNLKHLYKLVEECCTETVRMDLEKDKVITLDYKSAKKVTSKILKSLKIPSAVSEDL 469
Query: 408 K 406
+
Sbjct: 470 Q 470
>ref|NP_564856.1| DegP protease; protein id: At1g65630.1 [Arabidopsis thaliana]
gi|6686413|gb|AAF23847.1|AC007234_19 F1E22.1
[Arabidopsis thaliana]
Length = 559
Score = 50.4 bits (119), Expect = 2e-05
Identities = 27/61 (44%), Positives = 39/61 (63%)
Frame = -3
Query: 588 GKPVKNLKSLAAMVESCRDEYLKFNLEYDQIVVLRTKTAKAATLDILATHCIPSAMSDDL 409
G V NLK L +VE C E ++ +LE D+++ L K+AK T IL + IPSA+S+DL
Sbjct: 482 GVQVHNLKHLYKLVEECCTETVRMDLEKDKVITLDYKSAKKVTSKILKSLKIPSAVSEDL 541
Query: 408 K 406
+
Sbjct: 542 Q 542
>ref|NP_566552.1| DegP protease; protein id: At3g16550.1 [Arabidopsis thaliana]
Length = 491
Score = 50.1 bits (118), Expect = 2e-05
Identities = 25/59 (42%), Positives = 38/59 (64%)
Frame = -3
Query: 588 GKPVKNLKSLAAMVESCRDEYLKFNLEYDQIVVLRTKTAKAATLDILATHCIPSAMSDD 412
G VKNLK L ++E C + L+ +LE D+++VL ++AK AT +IL H I SA + +
Sbjct: 433 GVKVKNLKHLRELIEGCFSKDLRLDLENDKVMVLNYESAKKATFEILERHNIKSAWASE 491
>dbj|BAB01154.1| contains similarity to serine protease~gene_id:MDC8.18 [Arabidopsis
thaliana]
Length = 499
Score = 50.1 bits (118), Expect = 2e-05
Identities = 25/59 (42%), Positives = 38/59 (64%)
Frame = -3
Query: 588 GKPVKNLKSLAAMVESCRDEYLKFNLEYDQIVVLRTKTAKAATLDILATHCIPSAMSDD 412
G VKNLK L ++E C + L+ +LE D+++VL ++AK AT +IL H I SA + +
Sbjct: 441 GVKVKNLKHLRELIEGCFSKDLRLDLENDKVMVLNYESAKKATFEILERHNIKSAWASE 499
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 471,545,416
Number of Sequences: 1393205
Number of extensions: 9465455
Number of successful extensions: 18747
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 18342
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18739
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)