Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
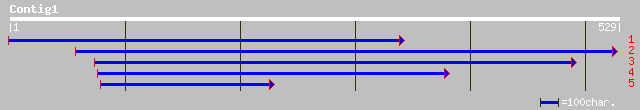
Query= KMC002419A_C01 KMC002419A_c01
(527 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T09591 probable cdc2-like protein kinase cdc2MsF - alfalfa ... 134 7e-31
emb|CAC15504.1| B2-type cyclin dependent kinase [Lycopersicon es... 131 4e-30
gb|AAM61014.1| putative cell division control protein cdc2 kinas... 127 8e-29
ref|NP_173517.1| Cyclin-dependent kinase B2;2; protein id: At1g2... 127 8e-29
sp|Q38775|CC2D_ANTMA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG D g... 123 1e-27
>pir||T09591 probable cdc2-like protein kinase cdc2MsF - alfalfa
gi|1806146|emb|CAA65982.1| cdc2MsF [Medicago sativa]
Length = 316
Score = 134 bits (337), Expect = 7e-31
Identities = 60/72 (83%), Positives = 66/72 (91%)
Frame = -3
Query: 525 PNEEVWPGVSKLMNWHEYPQWNPQSLSTAVPNLDELGLDLLSEMLHYEPSKRISAKKAME 346
PNE+VWPGVSKLMNWHEYPQW PQSLS AVP L+E G+DLLS+ML YEPSKR+SAKKAME
Sbjct: 245 PNEDVWPGVSKLMNWHEYPQWGPQSLSKAVPGLEEAGVDLLSQMLQYEPSKRLSAKKAME 304
Query: 345 HCYFDDLDKTYL 310
H YFDDLDKT+L
Sbjct: 305 HPYFDDLDKTHL 316
>emb|CAC15504.1| B2-type cyclin dependent kinase [Lycopersicon esculentum]
Length = 315
Score = 131 bits (330), Expect = 4e-30
Identities = 60/72 (83%), Positives = 64/72 (88%)
Frame = -3
Query: 525 PNEEVWPGVSKLMNWHEYPQWNPQSLSTAVPNLDELGLDLLSEMLHYEPSKRISAKKAME 346
PNEE+WPGVSKL+NWHEYPQW PQ LST VP LDE G+ LLSEMLHYEPS+RISAKKAME
Sbjct: 244 PNEELWPGVSKLVNWHEYPQWKPQPLSTVVPGLDEDGIHLLSEMLHYEPSRRISAKKAME 303
Query: 345 HCYFDDLDKTYL 310
H YFDDLDKT L
Sbjct: 304 HPYFDDLDKTPL 315
>gb|AAM61014.1| putative cell division control protein cdc2 kinase [Arabidopsis
thaliana]
Length = 303
Score = 127 bits (319), Expect = 8e-29
Identities = 61/73 (83%), Positives = 65/73 (88%), Gaps = 1/73 (1%)
Frame = -3
Query: 525 PNEEVWPGVSKLMNWHEYPQWNPQSLSTAVPNLDELGLDLLSEMLHYEPSKRISAKKAME 346
PNEEVWPGVSKL +WHEYPQW P SLSTAVPNLDE GLDLLS+ML YEP+KRISAKKAME
Sbjct: 231 PNEEVWPGVSKLKDWHEYPQWKPLSLSTAVPNLDEAGLDLLSKMLEYEPAKRISAKKAME 290
Query: 345 HCYFDDL-DKTYL 310
H YFDDL DK+ L
Sbjct: 291 HPYFDDLPDKSSL 303
>ref|NP_173517.1| Cyclin-dependent kinase B2;2; protein id: At1g20930.1, supported by
cDNA: 108339. [Arabidopsis thaliana]
gi|25287586|pir||B86342 probable cdc2 kinase [imported]
- Arabidopsis thaliana
gi|4836894|gb|AAD30597.1|AC007369_7 Putative cdc2 kinase
[Arabidopsis thaliana]
Length = 315
Score = 127 bits (319), Expect = 8e-29
Identities = 61/73 (83%), Positives = 65/73 (88%), Gaps = 1/73 (1%)
Frame = -3
Query: 525 PNEEVWPGVSKLMNWHEYPQWNPQSLSTAVPNLDELGLDLLSEMLHYEPSKRISAKKAME 346
PNEEVWPGVSKL +WHEYPQW P SLSTAVPNLDE GLDLLS+ML YEP+KRISAKKAME
Sbjct: 243 PNEEVWPGVSKLKDWHEYPQWKPLSLSTAVPNLDEAGLDLLSKMLEYEPAKRISAKKAME 302
Query: 345 HCYFDDL-DKTYL 310
H YFDDL DK+ L
Sbjct: 303 HPYFDDLPDKSSL 315
>sp|Q38775|CC2D_ANTMA CELL DIVISION CONTROL PROTEIN 2 HOMOLOG D gi|7489308|pir||T17118
protein kinase cdc2d (EC 2.7.1.-), cyclin-dependent -
garden snapdragon gi|1321678|emb|CAA66236.1|
cyclin-dependent kinase [Antirrhinum majus]
Length = 312
Score = 123 bits (309), Expect = 1e-27
Identities = 55/72 (76%), Positives = 64/72 (88%)
Frame = -3
Query: 525 PNEEVWPGVSKLMNWHEYPQWNPQSLSTAVPNLDELGLDLLSEMLHYEPSKRISAKKAME 346
PNEE+WPGVS L++WHEYPQW Q +S+AVP LDE GL+LLSEMLHYEPS+RISAKKAME
Sbjct: 241 PNEEIWPGVSTLVDWHEYPQWTAQPISSAVPGLDEKGLNLLSEMLHYEPSRRISAKKAME 300
Query: 345 HCYFDDLDKTYL 310
H YFD+LDK+ L
Sbjct: 301 HPYFDELDKSGL 312
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 417,617,795
Number of Sequences: 1393205
Number of extensions: 8178998
Number of successful extensions: 21218
Number of sequences better than 10.0: 1014
Number of HSP's better than 10.0 without gapping: 20348
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20931
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17308240320
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)