Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002407A_C01 KMC002407A_c01
(572 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
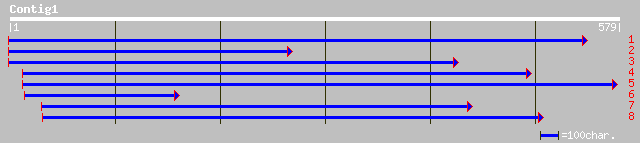
gb|AAC32134.1| KIAA0107-like protein [Picea mariana] 233 1e-60
ref|NP_567709.1| 26S proteasome regulatory subunit (RPN7), putat... 233 2e-60
sp|Q8W425|PSD6_ORYSA 26S proteasome non-ATPase regulatory subuni... 232 3e-60
pir||T06666 26S proteasome regulatory particle chain RPN7 homolo... 220 1e-56
emb|CAC09486.1| contains similarity to F6I7.30 [Oryza sativa (in... 211 7e-54
>gb|AAC32134.1| KIAA0107-like protein [Picea mariana]
Length = 232
Score = 233 bits (595), Expect = 1e-60
Identities = 114/130 (87%), Positives = 122/130 (93%)
Frame = -1
Query: 572 VNSVYDCQYKSLFSAFAGLTQQIKLDRYLHPHFRYYMREIRTVVYSQFLESYKSVTIEAM 393
+NS+Y CQYKS F AFAGLT QI+LDRYLHPHFRYYMRE+RTVVYSQFLESYKSVT+EAM
Sbjct: 103 LNSLYGCQYKSFFVAFAGLTDQIRLDRYLHPHFRYYMREVRTVVYSQFLESYKSVTMEAM 162
Query: 392 AKAFGVTVDFIDVELSRFIAAGKLHCKIDKFAGVLETNRPDAKNALYQTTIKQGDFLLNR 213
A+AFGV+VDFID ELSRFIAAG+LHCKIDK GVLETNRPDAKNALYQ TIKQGDFLLNR
Sbjct: 163 ARAFGVSVDFIDRELSRFIAAGRLHCKIDKVVGVLETNRPDAKNALYQATIKQGDFLLNR 222
Query: 212 IQKLSRVIDL 183
IQKLSRVIDL
Sbjct: 223 IQKLSRVIDL 232
>ref|NP_567709.1| 26S proteasome regulatory subunit (RPN7), putative; protein id:
At4g24820.1, supported by cDNA: 38927., supported by
cDNA: gi_15450839, supported by cDNA: gi_20259841
[Arabidopsis thaliana] gi|20978551|sp|Q93Y35|PSD6_ARATH
Probable 26S proteasome non-ATPase regulatory subunit 6
gi|15450840|gb|AAK96691.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
gi|20259842|gb|AAM13268.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
gi|21593433|gb|AAM65400.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
gi|23397029|gb|AAN31800.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
Length = 387
Score = 233 bits (593), Expect = 2e-60
Identities = 113/130 (86%), Positives = 123/130 (93%)
Frame = -1
Query: 572 VNSVYDCQYKSLFSAFAGLTQQIKLDRYLHPHFRYYMREIRTVVYSQFLESYKSVTIEAM 393
+NS+Y+CQYK+ FSAFAG+ QIK DRYL+PHFR+YMRE+RTVVYSQFLESYKSVT+EAM
Sbjct: 258 LNSLYECQYKAFFSAFAGMAVQIKYDRYLYPHFRFYMREVRTVVYSQFLESYKSVTVEAM 317
Query: 392 AKAFGVTVDFIDVELSRFIAAGKLHCKIDKFAGVLETNRPDAKNALYQTTIKQGDFLLNR 213
AKAFGV+VDFID ELSRFIAAGKLHCKIDK AGVLETNRPDAKNALYQ TIKQGDFLLNR
Sbjct: 318 AKAFGVSVDFIDQELSRFIAAGKLHCKIDKVAGVLETNRPDAKNALYQATIKQGDFLLNR 377
Query: 212 IQKLSRVIDL 183
IQKLSRVIDL
Sbjct: 378 IQKLSRVIDL 387
>sp|Q8W425|PSD6_ORYSA 26S proteasome non-ATPase regulatory subunit 6 (26S proteasome
regulatory particle non-ATPase subunit 7) (OsRPN7)
gi|17297977|dbj|BAB78486.1| 26S proteasome regulatory
particle non-ATPase subunit7 [Oryza sativa (japonica
cultivar-group)] gi|21741449|emb|CAD41393.1|
OJ000223_09.5 [Oryza sativa (japonica cultivar-group)]
Length = 389
Score = 232 bits (591), Expect = 3e-60
Identities = 113/130 (86%), Positives = 123/130 (93%)
Frame = -1
Query: 572 VNSVYDCQYKSLFSAFAGLTQQIKLDRYLHPHFRYYMREIRTVVYSQFLESYKSVTIEAM 393
+NS+Y+CQYKS F+AF+GLT+QIKLDRYL PHFRYYMRE+RTVVYSQFLESYKSVT+EAM
Sbjct: 260 LNSLYNCQYKSFFAAFSGLTEQIKLDRYLQPHFRYYMREVRTVVYSQFLESYKSVTMEAM 319
Query: 392 AKAFGVTVDFIDVELSRFIAAGKLHCKIDKFAGVLETNRPDAKNALYQTTIKQGDFLLNR 213
A AFGVTVDFID+ELSRFIAAGKLHCKIDK A VLETNRPDA+NA YQ TIKQGDFLLNR
Sbjct: 320 ASAFGVTVDFIDLELSRFIAAGKLHCKIDKVACVLETNRPDARNAFYQATIKQGDFLLNR 379
Query: 212 IQKLSRVIDL 183
IQKLSRVIDL
Sbjct: 380 IQKLSRVIDL 389
>pir||T06666 26S proteasome regulatory particle chain RPN7 homolog F6I7.30 -
Arabidopsis thaliana gi|4678261|emb|CAB41122.1| putative
proteasome regulatory subunit [Arabidopsis thaliana]
gi|7269333|emb|CAB79392.1| putative proteasome
regulatory subunit [Arabidopsis thaliana]
Length = 406
Score = 220 bits (560), Expect = 1e-56
Identities = 112/149 (75%), Positives = 123/149 (82%), Gaps = 19/149 (12%)
Frame = -1
Query: 572 VNSVYDCQYKSLFSAF-------------------AGLTQQIKLDRYLHPHFRYYMREIR 450
+NS+Y+CQYK+ FSAF +G+ QIK DRYL+PHFR+YMRE+R
Sbjct: 258 LNSLYECQYKAFFSAFGKFITRTRGHTSECMLLGFSGMAVQIKYDRYLYPHFRFYMREVR 317
Query: 449 TVVYSQFLESYKSVTIEAMAKAFGVTVDFIDVELSRFIAAGKLHCKIDKFAGVLETNRPD 270
TVVYSQFLESYKSVT+EAMAKAFGV+VDFID ELSRFIAAGKLHCKIDK AGVLETNRPD
Sbjct: 318 TVVYSQFLESYKSVTVEAMAKAFGVSVDFIDQELSRFIAAGKLHCKIDKVAGVLETNRPD 377
Query: 269 AKNALYQTTIKQGDFLLNRIQKLSRVIDL 183
AKNALYQ TIKQGDFLLNRIQKLSRVIDL
Sbjct: 378 AKNALYQATIKQGDFLLNRIQKLSRVIDL 406
>emb|CAC09486.1| contains similarity to F6I7.30 [Oryza sativa (indica
cultivar-group)]
Length = 209
Score = 211 bits (536), Expect = 7e-54
Identities = 104/114 (91%), Positives = 109/114 (95%)
Frame = -1
Query: 524 AGLTQQIKLDRYLHPHFRYYMREIRTVVYSQFLESYKSVTIEAMAKAFGVTVDFIDVELS 345
AGLT+QIKLDRYL PHFRYYMRE+RTVVYSQFLESYKSVT+EAMA AFGVTVDFID+ELS
Sbjct: 96 AGLTEQIKLDRYLQPHFRYYMREVRTVVYSQFLESYKSVTMEAMASAFGVTVDFIDLELS 155
Query: 344 RFIAAGKLHCKIDKFAGVLETNRPDAKNALYQTTIKQGDFLLNRIQKLSRVIDL 183
RFIAAGKLHCKIDK AGVLETNRPDA+NA YQ TIKQGDFLLNRIQKLSRVIDL
Sbjct: 156 RFIAAGKLHCKIDKVAGVLETNRPDARNAFYQATIKQGDFLLNRIQKLSRVIDL 209
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 444,399,174
Number of Sequences: 1393205
Number of extensions: 8792561
Number of successful extensions: 18884
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 18488
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18873
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)