Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
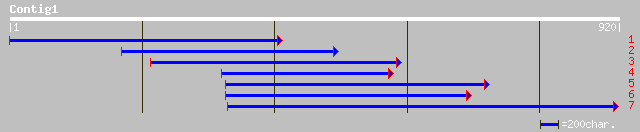
Query= KMC002395A_C01 KMC002395A_c01
(907 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_563676.1| expressed protein; protein id: At1g03140.1, sup... 142 8e-58
gb|AAL83635.1|AC093017_19 putative potassium channel regulatory ... 135 5e-56
ref|NP_175861.1| potassium channel regulatory factor, putative; ... 125 3e-51
ref|NP_612532.1| potassium channel regulatory factor [Rattus nor... 103 2e-29
ref|NP_003666.1| PRP18 pre-mRNA processing factor 18 homolog; pr... 103 3e-29
>ref|NP_563676.1| expressed protein; protein id: At1g03140.1, supported by cDNA:
gi_15293172 [Arabidopsis thaliana]
gi|25518006|pir||E86162 hypothetical protein F10O3.3 -
Arabidopsis thaliana gi|4587563|gb|AAD25794.1|AC006550_2
Similar to gb|U51990 pre-mRNA-splicing factor hPrp18
from Homo sapiens. ESTs gb|T46391 and gb|AA721815 come
from this gene. [Arabidopsis thaliana]
gi|13430636|gb|AAK25940.1|AF360230_1 unknown protein
[Arabidopsis thaliana] gi|15293173|gb|AAK93697.1|
unknown protein [Arabidopsis thaliana]
Length = 420
Score = 142 bits (359), Expect(2) = 8e-58
Identities = 71/81 (87%), Positives = 76/81 (93%)
Frame = -2
Query: 657 LGNAPWPIGVTMVGIHERSAREKIHTNSVAHIMNDETTRKYLQSVKRLMSICQLLYPTLP 478
+GNAPWPIGVTMVGIHERSAREKI+TNSVAHIMNDETTRKYLQSVKRLM+ CQ YPT+P
Sbjct: 315 IGNAPWPIGVTMVGIHERSAREKIYTNSVAHIMNDETTRKYLQSVKRLMTFCQRRYPTMP 374
Query: 477 SKAFEFNSLANGGSDLKSLLA 415
SKA EFNSLAN GSDL+SLLA
Sbjct: 375 SKAVEFNSLAN-GSDLQSLLA 394
Score = 104 bits (259), Expect(2) = 8e-58
Identities = 55/88 (62%), Positives = 62/88 (69%)
Frame = -1
Query: 907 FKKLLNEWKVQLLREMPEAKRRTADGKSLVARFKQCARYLNPLFKFCRNKILPDDIRQAL 728
+KKLL EWK Q L M +RRTA GK +VA FKQCARYL PLF CR K LP DIRQAL
Sbjct: 233 YKKLLIEWK-QELDAMENTERRTAKGKQMVATFKQCARYLVPLFNLCRKKGLPADIRQAL 291
Query: 727 LLMVECFMKRDYQAAMSHYMKLAARECP 644
++MV +KRDY AAM HY+KLA P
Sbjct: 292 MVMVNHCIKRDYLAAMDHYIKLAIGNAP 319
>gb|AAL83635.1|AC093017_19 putative potassium channel regulatory factor [Oryza sativa
(japonica cultivar-group)]
Length = 501
Score = 135 bits (339), Expect(2) = 5e-56
Identities = 67/81 (82%), Positives = 74/81 (90%)
Frame = -2
Query: 657 LGNAPWPIGVTMVGIHERSAREKIHTNSVAHIMNDETTRKYLQSVKRLMSICQLLYPTLP 478
+GN+PWPIGVTMVGIHERSAREKI+ NSVAHIMNDETTRKYLQSVKRLM+ CQ YPT P
Sbjct: 405 IGNSPWPIGVTMVGIHERSAREKIYANSVAHIMNDETTRKYLQSVKRLMTFCQRKYPTDP 464
Query: 477 SKAFEFNSLANGGSDLKSLLA 415
S++ EFNSLAN GSDL+SLLA
Sbjct: 465 SRSVEFNSLAN-GSDLQSLLA 484
Score = 105 bits (263), Expect(2) = 5e-56
Identities = 51/88 (57%), Positives = 66/88 (74%)
Frame = -1
Query: 907 FKKLLNEWKVQLLREMPEAKRRTADGKSLVARFKQCARYLNPLFKFCRNKILPDDIRQAL 728
FK+L++EW Q + EMPEA+RRTA GK+ VA KQCARYL PLFK C+ K LP+D+R +L
Sbjct: 323 FKRLMSEWS-QEMDEMPEAERRTAKGKAAVATCKQCARYLEPLFKLCKKKTLPEDVRGSL 381
Query: 727 LLMVECFMKRDYQAAMSHYMKLAARECP 644
L +V C M+RDY AA+ +Y+KLA P
Sbjct: 382 LEVVRCCMRRDYLAAVDNYIKLAIGNSP 409
>ref|NP_175861.1| potassium channel regulatory factor, putative; protein id:
At1g54590.1 [Arabidopsis thaliana]
gi|25405750|pir||A96588 hypothetical protein T22H22.4
[imported] - Arabidopsis thaliana
gi|3776560|gb|AAC64877.1| Similar to gb|U51990 hPrp18
(splicing factor) gene from Homo sapiens. [Arabidopsis
thaliana]
Length = 256
Score = 125 bits (314), Expect(2) = 3e-51
Identities = 66/82 (80%), Positives = 72/82 (87%), Gaps = 1/82 (1%)
Frame = -2
Query: 657 LGNAPWPIGVTMVGIHERSAREKIHTNS-VAHIMNDETTRKYLQSVKRLMSICQLLYPTL 481
+GNAPWPIGVTMVGIHERSAREKI T+S VAHIMN+ETTRKYLQSVKRLM+ CQ Y L
Sbjct: 150 IGNAPWPIGVTMVGIHERSAREKISTSSSVAHIMNNETTRKYLQSVKRLMTFCQRRYSAL 209
Query: 480 PSKAFEFNSLANGGSDLKSLLA 415
PSK+ EFNSLAN GS+L SLLA
Sbjct: 210 PSKSIEFNSLAN-GSNLHSLLA 230
Score = 99.8 bits (247), Expect(2) = 3e-51
Identities = 50/87 (57%), Positives = 60/87 (68%)
Frame = -1
Query: 904 KKLLNEWKVQLLREMPEAKRRTADGKSLVARFKQCARYLNPLFKFCRNKILPDDIRQALL 725
KKLL EWK Q L M +RRTA GK ++A F QCARYL PLF CRNK LP DIRQ L+
Sbjct: 69 KKLLLEWK-QELEAMENTERRTAIGKQMLATFNQCARYLTPLFHLCRNKCLPADIRQGLM 127
Query: 724 LMVECFMKRDYQAAMSHYMKLAARECP 644
+MV C++KRDY A + ++KLA P
Sbjct: 128 VMVNCWIKRDYLDATAQFIKLAIGNAP 154
>ref|NP_612532.1| potassium channel regulatory factor [Rattus norvegicus]
gi|11360406|pir||JC7358 potassium channel regulatory
factor - rat gi|7271971|gb|AAF44715.1|AF244920_1
potassium channel regulatory factor [Rattus norvegicus]
Length = 342
Score = 103 bits (257), Expect(2) = 2e-29
Identities = 46/69 (66%), Positives = 57/69 (81%)
Frame = -2
Query: 657 LGNAPWPIGVTMVGIHERSAREKIHTNSVAHIMNDETTRKYLQSVKRLMSICQLLYPTLP 478
+GNAPWPIGVTMVGIH R+ REKI + VAH++NDET RKY+Q +KRLM+ICQ +PT P
Sbjct: 274 IGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYIQGLKRLMTICQKHFPTDP 333
Query: 477 SKAFEFNSL 451
SK E+N+L
Sbjct: 334 SKCVEYNAL 342
Score = 48.1 bits (113), Expect(2) = 2e-29
Identities = 27/87 (31%), Positives = 46/87 (52%)
Frame = -1
Query: 904 KKLLNEWKVQLLREMPEAKRRTADGKSLVARFKQCARYLNPLFKFCRNKILPDDIRQALL 725
K LL W +L + +R+ GK A KQ YL PLF+ R + LP DI++++
Sbjct: 193 KFLLGVWAKEL-NSREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADIKESIT 251
Query: 724 LMVECFMKRDYQAAMSHYMKLAARECP 644
+++ ++R+Y A Y+++A P
Sbjct: 252 DIIKFMLQREYVKANDAYLQMAIGNAP 278
>ref|NP_003666.1| PRP18 pre-mRNA processing factor 18 homolog; pre-mRNA splicing
factor similar to S. cerevisiae Prp18; pre-mRNA
processing factor 18 [Homo sapiens]
gi|1805249|gb|AAB41490.1| hPrp18
gi|12653993|gb|AAH00794.1|AAH00794 pre-mRNA processing
factor 18 [Homo sapiens]
Length = 342
Score = 103 bits (257), Expect(2) = 3e-29
Identities = 46/69 (66%), Positives = 57/69 (81%)
Frame = -2
Query: 657 LGNAPWPIGVTMVGIHERSAREKIHTNSVAHIMNDETTRKYLQSVKRLMSICQLLYPTLP 478
+GNAPWPIGVTMVGIH R+ REKI + VAH++NDET RKY+Q +KRLM+ICQ +PT P
Sbjct: 274 IGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYIQGLKRLMTICQKHFPTDP 333
Query: 477 SKAFEFNSL 451
SK E+N+L
Sbjct: 334 SKCVEYNAL 342
Score = 47.8 bits (112), Expect(2) = 3e-29
Identities = 27/87 (31%), Positives = 46/87 (52%)
Frame = -1
Query: 904 KKLLNEWKVQLLREMPEAKRRTADGKSLVARFKQCARYLNPLFKFCRNKILPDDIRQALL 725
K LL W +L + +R+ GK A KQ YL PLF+ R + LP DI++++
Sbjct: 193 KFLLGVWAKEL-NAREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADIKESIT 251
Query: 724 LMVECFMKRDYQAAMSHYMKLAARECP 644
+++ ++R+Y A Y+++A P
Sbjct: 252 DIIKFMLQREYVKANDAYLQMAIGNAP 278
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 735,407,646
Number of Sequences: 1393205
Number of extensions: 15212296
Number of successful extensions: 35299
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 34163
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35277
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 49363855696
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)