Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
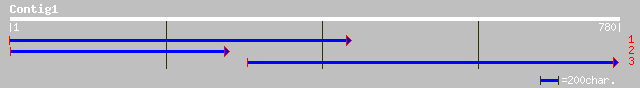
Query= KMC002389A_C01 KMC002389A_c01
(780 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_196661.1| putative protein; protein id: At5g11000.1, supp... 124 2e-27
ref|NP_565589.2| hypothetical protein; protein id: At2g25200.1 [... 80 4e-14
ref|NP_193005.1| putative protein; protein id: At4g12690.1, supp... 66 6e-10
pir||C84455 hypothetical protein At2g04220 [imported] - Arabidop... 65 8e-10
ref|NP_178505.1| hypothetical protein; protein id: At2g04220.1 [... 65 8e-10
>ref|NP_196661.1| putative protein; protein id: At5g11000.1, supported by cDNA:
263168., supported by cDNA: gi_16323144 [Arabidopsis
thaliana] gi|11358298|pir||T50809 hypothetical protein
T30N20_270 - Arabidopsis thaliana
gi|8979734|emb|CAB96855.1| putative protein [Arabidopsis
thaliana] gi|16323145|gb|AAL15307.1|
AT5g11000/T30N20_270 [Arabidopsis thaliana]
gi|21436003|gb|AAM51579.1| AT5g11000/T30N20_270
[Arabidopsis thaliana] gi|21554751|gb|AAM63680.1|
unknown [Arabidopsis thaliana]
Length = 389
Score = 124 bits (311), Expect = 2e-27
Identities = 79/171 (46%), Positives = 99/171 (57%), Gaps = 39/171 (22%)
Frame = -2
Query: 779 KVMQIKRLKWKFRGNERVEVDGVPIQISWDVYNWLFQKDNS------DGHAIFMFKFEEE 618
+V+QIKRL+WKFRGNE+VE+DGV +QISWDVYNWLFQ +S GHA+FMF+FE +
Sbjct: 217 QVLQIKRLRWKFRGNEKVEIDGVHVQISWDVYNWLFQSKSSGDGGGGGGHAVFMFRFESD 276
Query: 617 EEDE--------EDQRGKERNLVNLWTHQNL------NLGVSEWGKV--------GSSSS 504
E E E++ K RN + LW + G+ EW K+ SSSS
Sbjct: 277 PEAEEVCETKRKEEEDEKNRNGIVLWKPKKQCGNSFGIKGIVEWRKMRKRFVKSKRSSSS 336
Query: 503 ASMASSGGSFGGSSSVMEW-SSVEENEL----------VVPLGFSLLVYAW 384
+S++ S S SSSVMEW SS +E E LGFSLLVYAW
Sbjct: 337 SSISMSSASSACSSSVMEWASSADEAEYGGGGCSGSGSGNGLGFSLLVYAW 387
>ref|NP_565589.2| hypothetical protein; protein id: At2g25200.1 [Arabidopsis
thaliana] gi|25412287|pir||E84645 hypothetical protein
At2g25200 [imported] - Arabidopsis thaliana
gi|4567257|gb|AAD23671.1| hypothetical protein
[Arabidopsis thaliana]
Length = 354
Score = 79.7 bits (195), Expect = 4e-14
Identities = 52/141 (36%), Positives = 75/141 (52%), Gaps = 8/141 (5%)
Frame = -2
Query: 776 VMQIKRLKWKFRGNERVEVDGVPIQISWDVYNWLF---QKDNSDG-HAIFMFKFEEEEED 609
V++I +L+WKFRGN ++ +DGV IQISWDV+NWLF K D A+F+ +FE +E +
Sbjct: 223 VLKISQLQWKFRGNTKIVIDGVTIQISWDVFNWLFGGKDKVKPDKIPAVFLLRFENQEVE 282
Query: 608 EEDQRGKERNLVNLWTHQNLNLGV-SEWGKVGSSSSASMASSGGSFGGSSSVMEWSSV-- 438
D + + + + N S W S S+S SSVMEWSS
Sbjct: 283 GNDVLMMNKRVRDDVILRKENCRTPSFWASTYSHWSSSR---------MSSVMEWSSCRE 333
Query: 437 -EENELVVPLGFSLLVYAWKR 378
+E FSL++YAW++
Sbjct: 334 EDERSFGSKSWFSLIIYAWRK 354
>ref|NP_193005.1| putative protein; protein id: At4g12690.1, supported by cDNA:
12923. [Arabidopsis thaliana] gi|7459232|pir||T06627
hypothetical protein T20K18.40 - Arabidopsis thaliana
gi|4586245|emb|CAB40986.1| putative protein [Arabidopsis
thaliana] gi|7267970|emb|CAB78311.1| putative protein
[Arabidopsis thaliana] gi|21537307|gb|AAM61648.1|
unknown [Arabidopsis thaliana]
Length = 285
Score = 65.9 bits (159), Expect = 6e-10
Identities = 27/60 (45%), Positives = 42/60 (70%)
Frame = -2
Query: 776 VMQIKRLKWKFRGNERVEVDGVPIQISWDVYNWLFQKDNSDGHAIFMFKFEEEEEDEEDQ 597
++Q++ L+WKFRGN+ V VD P+Q+ WDVY+WLF + GH +F+FK E E + ++
Sbjct: 202 LVQVRNLQWKFRGNQTVLVDKEPVQVFWDVYDWLFSTPGT-GHGLFIFKPESGESETSNE 260
>pir||C84455 hypothetical protein At2g04220 [imported] - Arabidopsis thaliana
Length = 246
Score = 65.5 bits (158), Expect = 8e-10
Identities = 28/62 (45%), Positives = 42/62 (67%)
Frame = -2
Query: 776 VMQIKRLKWKFRGNERVEVDGVPIQISWDVYNWLFQKDNSDGHAIFMFKFEEEEEDEEDQ 597
++Q+K L+WKFRGN+ V VD P+Q+ WDVY+WLF + GH +F+FK E+ + +
Sbjct: 146 LIQVKNLQWKFRGNQTVLVDKQPVQVFWDVYDWLFSMPGT-GHGLFIFKPGTTEDSDMEG 204
Query: 596 RG 591
G
Sbjct: 205 SG 206
>ref|NP_178505.1| hypothetical protein; protein id: At2g04220.1 [Arabidopsis
thaliana] gi|20198211|gb|AAD27916.2| hypothetical
protein [Arabidopsis thaliana]
gi|20198251|gb|AAM15482.1| hypothetical protein
[Arabidopsis thaliana]
Length = 307
Score = 65.5 bits (158), Expect = 8e-10
Identities = 28/62 (45%), Positives = 42/62 (67%)
Frame = -2
Query: 776 VMQIKRLKWKFRGNERVEVDGVPIQISWDVYNWLFQKDNSDGHAIFMFKFEEEEEDEEDQ 597
++Q+K L+WKFRGN+ V VD P+Q+ WDVY+WLF + GH +F+FK E+ + +
Sbjct: 207 LIQVKNLQWKFRGNQTVLVDKQPVQVFWDVYDWLFSMPGT-GHGLFIFKPGTTEDSDMEG 265
Query: 596 RG 591
G
Sbjct: 266 SG 267
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 708,687,031
Number of Sequences: 1393205
Number of extensions: 17090948
Number of successful extensions: 84066
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 58884
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 77426
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38655378996
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)