Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002381A_C01 KMC002381A_c01
(802 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
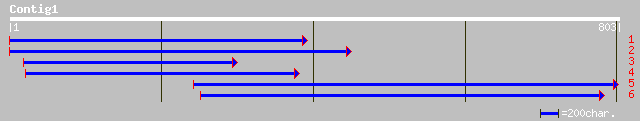
ref|NP_567245.1| bHLH protein; protein id: At4g02590.1, supporte... 183 3e-45
gb|AAM65759.1| putative lipoamide dehydrogenase [Arabidopsis tha... 181 8e-45
gb|AAM10948.1|AF488592_1 putative bHLH transcription factor [Ara... 181 1e-44
pir||B86161 F10O3.14 protein - Arabidopsis thaliana gi|4587574|g... 171 1e-41
ref|NP_563672.1| bHLH protein; protein id: At1g03040.1, supporte... 171 1e-41
>ref|NP_567245.1| bHLH protein; protein id: At4g02590.1, supported by cDNA: 4346.,
supported by cDNA: gi_13605858, supported by cDNA:
gi_20127057 [Arabidopsis thaliana]
gi|7486850|pir||T01090 hypothetical protein T10P11.13 -
Arabidopsis thaliana gi|3892050|gb|AAC78259.1|AAC78259
hypothetical protein [Arabidopsis thaliana]
gi|7269019|emb|CAB80752.1| hypothetical protein
[Arabidopsis thaliana]
gi|13605859|gb|AAK32915.1|AF367328_1 AT4g02590/T10P11_13
[Arabidopsis thaliana] gi|23506061|gb|AAN28890.1|
At4g02590/T10P11_13 [Arabidopsis thaliana]
Length = 310
Score = 183 bits (464), Expect = 3e-45
Identities = 96/131 (73%), Positives = 112/131 (85%), Gaps = 2/131 (1%)
Frame = -1
Query: 802 ELVPSINKSDRAAMLDEIVDYVKFLRLQVKVLSMSRLGGAGAVAQLVADVPLSA-VEGED 626
ELVP++NK+DRAAM+DEIVDYVKFLRLQVKVLSMSRLGGAGAVA LV D+PLS+ VE E
Sbjct: 177 ELVPTVNKTDRAAMIDEIVDYVKFLRLQVKVLSMSRLGGAGAVAPLVTDMPLSSSVEDET 236
Query: 625 IEGGASEQ-AWSKWSNDGTEQQVAKLMEEDVGAAMQFLQSKALCIMPISLASAIFRMPQS 449
EGG + Q AW KWSNDGTE+QVAKLMEE+VGAAMQ LQSKALC+MPISLA AI+
Sbjct: 237 GEGGRTPQPAWEKWSNDGTERQVAKLMEENVGAAMQLLQSKALCMMPISLAMAIYHSQPP 296
Query: 448 EASTTIKPESD 416
+ S+ +KPE++
Sbjct: 297 DTSSVVKPENN 307
>gb|AAM65759.1| putative lipoamide dehydrogenase [Arabidopsis thaliana]
Length = 310
Score = 181 bits (460), Expect = 8e-45
Identities = 95/131 (72%), Positives = 111/131 (84%), Gaps = 2/131 (1%)
Frame = -1
Query: 802 ELVPSINKSDRAAMLDEIVDYVKFLRLQVKVLSMSRLGGAGAVAQLVADVPLSA-VEGED 626
ELVP++NK+DRAAM+DEIVDYVKFLRLQVKVLSMSRLGG GAVA LV D+PLS+ VE E
Sbjct: 177 ELVPTVNKTDRAAMIDEIVDYVKFLRLQVKVLSMSRLGGVGAVAPLVTDMPLSSSVEDET 236
Query: 625 IEGGASEQ-AWSKWSNDGTEQQVAKLMEEDVGAAMQFLQSKALCIMPISLASAIFRMPQS 449
EGG + Q AW KWSNDGTE+QVAKLMEE+VGAAMQ LQSKALC+MPISLA AI+
Sbjct: 237 GEGGRTPQPAWEKWSNDGTERQVAKLMEENVGAAMQLLQSKALCMMPISLAMAIYHSQPP 296
Query: 448 EASTTIKPESD 416
+ S+ +KPE++
Sbjct: 297 DTSSVVKPENN 307
>gb|AAM10948.1|AF488592_1 putative bHLH transcription factor [Arabidopsis thaliana]
Length = 310
Score = 181 bits (458), Expect = 1e-44
Identities = 95/131 (72%), Positives = 111/131 (84%), Gaps = 2/131 (1%)
Frame = -1
Query: 802 ELVPSINKSDRAAMLDEIVDYVKFLRLQVKVLSMSRLGGAGAVAQLVADVPLSA-VEGED 626
ELVP++NK+DRAAM+DEIVDYVKFLRLQVKVLSMSRLGGAGAVA LV D+PLS+ V E
Sbjct: 177 ELVPTVNKTDRAAMIDEIVDYVKFLRLQVKVLSMSRLGGAGAVAPLVTDMPLSSSVXDET 236
Query: 625 IEGGASEQ-AWSKWSNDGTEQQVAKLMEEDVGAAMQFLQSKALCIMPISLASAIFRMPQS 449
EGG + Q AW KWSNDGTE+QVAKLMEE+VGAAMQ LQSKALC+MPISLA AI+
Sbjct: 237 GEGGRTPQPAWEKWSNDGTERQVAKLMEENVGAAMQLLQSKALCMMPISLAMAIYHSQPP 296
Query: 448 EASTTIKPESD 416
+ S+ +KPE++
Sbjct: 297 DTSSVVKPENN 307
>pir||B86161 F10O3.14 protein - Arabidopsis thaliana
gi|4587574|gb|AAD25805.1|AC006550_13 Contains PF|00010
helix-loop-helix DNA-binding domain. ESTs gb|T45640 and
gb|T22783 come from this gene. [Arabidopsis thaliana]
Length = 297
Score = 171 bits (433), Expect = 1e-41
Identities = 92/129 (71%), Positives = 107/129 (82%), Gaps = 2/129 (1%)
Frame = -1
Query: 802 ELVPSINKSDRAAMLDEIVDYVKFLRLQVKVLSMSRLGGAGAVAQLVADVPL-SAVEGED 626
ELVP++NK+DRAAM+DEIVDYVKFLRLQVKVLSMSRLGGAGAVA LV ++PL S+VE E
Sbjct: 170 ELVPTVNKTDRAAMIDEIVDYVKFLRLQVKVLSMSRLGGAGAVAPLVTEMPLSSSVEDE- 228
Query: 625 IEGGASEQAWSKWSNDGTEQQVAKLMEEDVGAAMQFLQSKALCIMPISLASAIFR-MPQS 449
++ W KWSNDGTE+QVAKLMEE+VGAAMQ LQSKALCIMPISLA AI+ P
Sbjct: 229 -----TQAVWEKWSNDGTERQVAKLMEENVGAAMQLLQSKALCIMPISLAMAIYHSQPPD 283
Query: 448 EASTTIKPE 422
+S+ +KPE
Sbjct: 284 TSSSIVKPE 292
>ref|NP_563672.1| bHLH protein; protein id: At1g03040.1, supported by cDNA:
gi_15450778 [Arabidopsis thaliana]
gi|15450779|gb|AAK96661.1| Unknown protein [Arabidopsis
thaliana] gi|21387097|gb|AAM47952.1| unknown protein
[Arabidopsis thaliana]
gi|21735477|gb|AAL55714.2|AF251692_1 putative
transcription factor BHLH7 [Arabidopsis thaliana]
Length = 302
Score = 171 bits (433), Expect = 1e-41
Identities = 92/129 (71%), Positives = 107/129 (82%), Gaps = 2/129 (1%)
Frame = -1
Query: 802 ELVPSINKSDRAAMLDEIVDYVKFLRLQVKVLSMSRLGGAGAVAQLVADVPL-SAVEGED 626
ELVP++NK+DRAAM+DEIVDYVKFLRLQVKVLSMSRLGGAGAVA LV ++PL S+VE E
Sbjct: 175 ELVPTVNKTDRAAMIDEIVDYVKFLRLQVKVLSMSRLGGAGAVAPLVTEMPLSSSVEDE- 233
Query: 625 IEGGASEQAWSKWSNDGTEQQVAKLMEEDVGAAMQFLQSKALCIMPISLASAIFR-MPQS 449
++ W KWSNDGTE+QVAKLMEE+VGAAMQ LQSKALCIMPISLA AI+ P
Sbjct: 234 -----TQAVWEKWSNDGTERQVAKLMEENVGAAMQLLQSKALCIMPISLAMAIYHSQPPD 288
Query: 448 EASTTIKPE 422
+S+ +KPE
Sbjct: 289 TSSSIVKPE 297
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 663,411,018
Number of Sequences: 1393205
Number of extensions: 14247374
Number of successful extensions: 50835
Number of sequences better than 10.0: 160
Number of HSP's better than 10.0 without gapping: 48545
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 50715
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40616159090
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)