Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002368A_C01 KMC002368A_c01
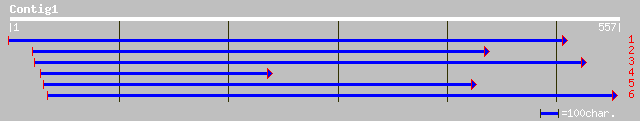
(557 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_566347.1| expressed protein; protein id: At3g09250.1, sup... 233 9e-61
dbj|BAB89544.1| P0435B05.23 [Oryza sativa (japonica cultivar-gro... 103 2e-21
ref|ZP_00073304.1| hypothetical protein [Trichodesmium erythraeu... 81 1e-14
ref|NP_567369.1| Expressed protein; protein id: At4g10925.1, sup... 74 9e-13
ref|NP_773477.1| blr6837 [Bradyrhizobium japonicum] gi|27355118|... 71 8e-12
>ref|NP_566347.1| expressed protein; protein id: At3g09250.1, supported by cDNA:
32811., supported by cDNA: gi_17380785, supported by
cDNA: gi_20259606 [Arabidopsis thaliana]
gi|6478918|gb|AAF14023.1|AC011436_7 unknown protein
[Arabidopsis thaliana] gi|17380786|gb|AAL36223.1|
unknown protein [Arabidopsis thaliana]
gi|20259607|gb|AAM14160.1| unknown protein [Arabidopsis
thaliana] gi|21592811|gb|AAM64760.1| unknown
[Arabidopsis thaliana]
Length = 244
Score = 233 bits (595), Expect = 9e-61
Identities = 104/160 (65%), Positives = 133/160 (83%), Gaps = 1/160 (0%)
Frame = -1
Query: 557 VDETTLVQELESAIAEENYAKAAEIRDILKNLKKDRETTLFGANSRFYESFKNGDLAAMQ 378
+DE TL Q+LE+A+ EENY +AA+IRD LK L++D + ++ ANSRFY+SF+NGDLAAMQ
Sbjct: 85 MDENTLKQDLETAVQEENYVEAAKIRDKLKELQEDNKASVLSANSRFYQSFRNGDLAAMQ 144
Query: 377 AMWAQSDEVCCVHPGMKGISGYDDVIESWNIVWANYEFPLEIKLEDIKFHVGEDMGYVTC 198
++W++S CCVHPG KGI+GYD V+ESW +VW NYEFPL I+L+D++ HV ++GYVTC
Sbjct: 145 SLWSKSGNPCCVHPGAKGITGYDYVMESWELVWMNYEFPLLIELKDVEVHVRGEVGYVTC 204
Query: 197 MEFVKTKG-GRWGGPFVTNVFERIDGPWFICVHHASPVDV 81
MEFVKTKG WG FV+NVFERIDG WFIC+HHASPVD+
Sbjct: 205 MEFVKTKGSSSWGAQFVSNVFERIDGQWFICIHHASPVDI 244
>dbj|BAB89544.1| P0435B05.23 [Oryza sativa (japonica cultivar-group)]
gi|20160737|dbj|BAB89678.1| P0503C12.2 [Oryza sativa
(japonica cultivar-group)]
Length = 377
Score = 103 bits (256), Expect = 2e-21
Identities = 47/91 (51%), Positives = 68/91 (74%)
Frame = -1
Query: 557 VDETTLVQELESAIAEENYAKAAEIRDILKNLKKDRETTLFGANSRFYESFKNGDLAAMQ 378
+DE TL + L++AI EE+Y++AA+IRD L+ L +D + +L AN+RFY +FKNGDLAAM
Sbjct: 76 IDEETLQRNLQTAIQEEDYSRAAKIRDDLRILHEDTKASLLAANTRFYNAFKNGDLAAMY 135
Query: 377 AMWAQSDEVCCVHPGMKGISGYDDVIESWNI 285
++WA+ D V +HP ISGYD V++SW +
Sbjct: 136 SLWAKGDHVYVIHPAAGRISGYDVVMQSWEM 166
>ref|ZP_00073304.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 128
Score = 80.9 bits (198), Expect = 1e-14
Identities = 41/133 (30%), Positives = 69/133 (51%), Gaps = 9/133 (6%)
Frame = -1
Query: 458 KDRETTLFGANSRFYESFKNGDLAAMQAMWAQSDEVCCVHPGMKGISGYDDVIESWNIVW 279
KD + N FY +F+ D+ A+ + +Q C+HPG I G++++ SW++++
Sbjct: 2 KDENQEILEVNQAFYRAFEKRDIIALHGILSQGISTVCIHPGRGAICGFENIRNSWDLIF 61
Query: 278 ANYEFPLEIKLEDIKFHVGEDMGYVTCME---------FVKTKGGRWGGPFVTNVFERID 126
N ++ +EI + I V D+GYV +E +K K TN+FE++
Sbjct: 62 KNTDY-IEIDTDVIIAEVNGDIGYVILVENIMQISRDTTIKDKS------IATNIFEKMG 114
Query: 125 GPWFICVHHASPV 87
G W++ HHASPV
Sbjct: 115 GNWYLINHHASPV 127
>ref|NP_567369.1| Expressed protein; protein id: At4g10925.1, supported by cDNA:
11425., supported by cDNA: gi_16604596, supported by
cDNA: gi_20466026 [Arabidopsis thaliana]
gi|16604597|gb|AAL24155.1| unknown protein [Arabidopsis
thaliana] gi|20466027|gb|AAM20348.1| unknown protein
[Arabidopsis thaliana] gi|21536887|gb|AAM61219.1|
unknown [Arabidopsis thaliana]
gi|26450596|dbj|BAC42410.1| unknown protein [Arabidopsis
thaliana]
Length = 231
Score = 74.3 bits (181), Expect = 9e-13
Identities = 38/117 (32%), Positives = 65/117 (55%), Gaps = 1/117 (0%)
Frame = -1
Query: 440 LFGANSRFYESFKNGDLAAMQAMWAQSDEVCCVHPGMKGISGYDDVIESWNIVWANYEFP 261
+ N+ F+ ++ L AM +W SD V C+H + SGY++VI+SW + + N+E
Sbjct: 108 IISVNTEFFTIIRDRALQAMARLWLNSDYVKCIHASGELFSGYNEVIQSWQLCF-NWEQG 166
Query: 260 LEIKLEDIKFHVGEDMGYVTCMEFVKTKGGRWGGPF-VTNVFERIDGPWFICVHHAS 93
+ ++ ++ + DM +VT ++ GGPF +TNVFE +G W + HH+S
Sbjct: 167 FDFQVHTVRTRILTDMAWVTMKAYLNVD----GGPFLITNVFEFHNGRWHMVHHHSS 219
>ref|NP_773477.1| blr6837 [Bradyrhizobium japonicum] gi|27355118|dbj|BAC52102.1|
blr6837 [Bradyrhizobium japonicum USDA 110]
Length = 135
Score = 71.2 bits (173), Expect = 8e-12
Identities = 39/122 (31%), Positives = 58/122 (46%)
Frame = -1
Query: 452 RETTLFGANSRFYESFKNGDLAAMQAMWAQSDEVCCVHPGMKGISGYDDVIESWNIVWAN 273
+++ + AN+ FY +F GD A M+ MWA D + C+HPG I G VI SW + N
Sbjct: 3 KDSKVIAANAAFYAAFSTGDFAGMEQMWADDDAISCIHPGWPAIIGRATVIGSWRDILQN 62
Query: 272 YEFPLEIKLEDIKFHVGEDMGYVTCMEFVKTKGGRWGGPFVTNVFERIDGPWFICVHHAS 93
E P +I + + V D V C+E + N F R+ W + H +S
Sbjct: 63 PERP-QIVCAEPQAIVDGDCARVLCIEILDGT-----ALAAANHFRRVGDDWRLVHHQSS 116
Query: 92 PV 87
P+
Sbjct: 117 PI 118
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 481,640,245
Number of Sequences: 1393205
Number of extensions: 10529190
Number of successful extensions: 26569
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 25811
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26554
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)