Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002361A_C02 KMC002361A_c02
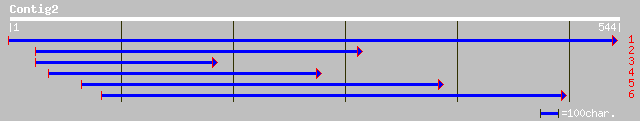
(521 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_175460.1| sterol delta7 reductase; protein id: At1g50430.... 179 2e-44
gb|AAC49278.1| sterol delta-7 reductase 170 1e-41
gb|AAL14611.1|AF417293_1 putative delta-7-sterol reductase [Cast... 136 1e-31
emb|CAB44317.1| lamin B receptor [Xenopus laevis] 110 1e-23
ref|NP_604448.1| lamin B receptor [Rattus norvegicus] gi|7513997... 108 5e-23
>ref|NP_175460.1| sterol delta7 reductase; protein id: At1g50430.1, supported by
cDNA: gi_1245181, supported by cDNA: gi_20466245
[Arabidopsis thaliana] gi|20140296|sp|Q9LDU6|ST7R_ARATH
7-dehydrocholesterol reductase (7-DHC reductase) (Sterol
delta-7-reductase) (Dwarf5 protein)
gi|25336689|pir||F96540 sterol delta7 reductase
[imported] - Arabidopsis thaliana
gi|7542561|gb|AAF63498.1|AF239701_1 sterol delta7
reductase [Arabidopsis thaliana]
gi|9454565|gb|AAF87888.1|AC012561_21 sterol delta7
reductase [Arabidopsis thaliana]
gi|20466246|gb|AAM20440.1| sterol delta7 reductase
[Arabidopsis thaliana] gi|23198074|gb|AAN15564.1| sterol
delta7 reductase [Arabidopsis thaliana]
Length = 432
Score = 179 bits (453), Expect = 2e-44
Identities = 79/89 (88%), Positives = 86/89 (95%)
Frame = -1
Query: 521 GQTKRSLLLTSGWWGLSRHFHYVPEILAAFFWTVPALFNHFLPYFYVIFLTILLFDRAKR 342
G+TK SLLLTSGWWGL+RHFHYVPEIL+AFFWTVPALF++FL YFYVIFLT+LLFDRAKR
Sbjct: 344 GETKTSLLLTSGWWGLARHFHYVPEILSAFFWTVPALFDNFLAYFYVIFLTLLLFDRAKR 403
Query: 341 DDDRCRSKYGKYWKLYCDKVPYRIIPGIY 255
DDDRCRSKYGKYWKLYC+KV YRIIPGIY
Sbjct: 404 DDDRCRSKYGKYWKLYCEKVKYRIIPGIY 432
>gb|AAC49278.1| sterol delta-7 reductase
Length = 430
Score = 170 bits (430), Expect = 1e-41
Identities = 77/89 (86%), Positives = 84/89 (93%)
Frame = -1
Query: 521 GQTKRSLLLTSGWWGLSRHFHYVPEILAAFFWTVPALFNHFLPYFYVIFLTILLFDRAKR 342
G+TK SLLLTSGWWGL+RHFHYVPEIL+AFFWTVPALF++FL YFYV LT+LLFDRAKR
Sbjct: 344 GETKTSLLLTSGWWGLARHFHYVPEILSAFFWTVPALFDNFLAYFYV--LTLLLFDRAKR 401
Query: 341 DDDRCRSKYGKYWKLYCDKVPYRIIPGIY 255
DDDRCRSKYGKYWKLYC+KV YRIIPGIY
Sbjct: 402 DDDRCRSKYGKYWKLYCEKVKYRIIPGIY 430
>gb|AAL14611.1|AF417293_1 putative delta-7-sterol reductase [Castanea sativa]
Length = 67
Score = 136 bits (343), Expect = 1e-31
Identities = 59/65 (90%), Positives = 65/65 (99%)
Frame = -1
Query: 449 EILAAFFWTVPALFNHFLPYFYVIFLTILLFDRAKRDDDRCRSKYGKYWKLYCDKVPYRI 270
+IL+AFFWTVPALFNHFLP+FYVIFLTILLFDRAKRDDDRCRSKYGKYWK++C+KVPYRI
Sbjct: 3 QILSAFFWTVPALFNHFLPFFYVIFLTILLFDRAKRDDDRCRSKYGKYWKVFCEKVPYRI 62
Query: 269 IPGIY 255
IPGIY
Sbjct: 63 IPGIY 67
>emb|CAB44317.1| lamin B receptor [Xenopus laevis]
Length = 620
Score = 110 bits (274), Expect = 1e-23
Identities = 48/84 (57%), Positives = 58/84 (68%)
Frame = -1
Query: 506 SLLLTSGWWGLSRHFHYVPEILAAFFWTVPALFNHFLPYFYVIFLTILLFDRAKRDDDRC 327
S LL SGWWG RH +Y+ +I+ A W + FNH LPYFYVIFLT+LL DRA RD+ RC
Sbjct: 537 SKLLISGWWGFVRHPNYLGDIIMALAWCLACGFNHILPYFYVIFLTLLLIDRAARDEQRC 596
Query: 326 RSKYGKYWKLYCDKVPYRIIPGIY 255
R KYG W YC V YR++P +Y
Sbjct: 597 REKYGLDWDKYCQHVRYRLLPYVY 620
>ref|NP_604448.1| lamin B receptor [Rattus norvegicus] gi|7513997|pir||JC5567 lamin B
receptor - rat gi|2204062|dbj|BAA20471.1| Rat NBP60
[Rattus norvegicus]
Length = 620
Score = 108 bits (269), Expect = 5e-23
Identities = 45/82 (54%), Positives = 57/82 (68%)
Frame = -1
Query: 500 LLTSGWWGLSRHFHYVPEILAAFFWTVPALFNHFLPYFYVIFLTILLFDRAKRDDDRCRS 321
LL SGWWG RH +Y+ +++ A W++P FNH LPYFYVI+ T LL R RD+ +CR
Sbjct: 539 LLVSGWWGFVRHPNYLGDLIMALAWSLPCGFNHILPYFYVIYFTALLIHREARDEHQCRR 598
Query: 320 KYGKYWKLYCDKVPYRIIPGIY 255
KYG W+ YC +VPYRI P IY
Sbjct: 599 KYGLAWEKYCQRVPYRIFPYIY 620
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 406,444,018
Number of Sequences: 1393205
Number of extensions: 7883124
Number of successful extensions: 18997
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 18635
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18993
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16731298976
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)