Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
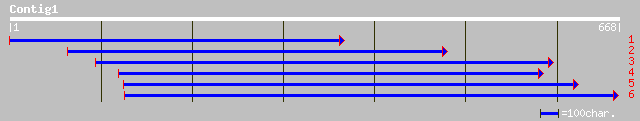
Query= KMC002355A_C01 KMC002355A_c01
(667 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG12330.1|AF249743_1 heterotrimeric GTP-binding protein subu... 207 1e-52
sp|P93398|GBB2_TOBAC Guanine nucleotide-binding protein beta sub... 206 2e-52
sp|P93397|GBB1_TOBAC Guanine nucleotide-binding protein beta sub... 204 1e-51
gb|AAL07488.1|AF414114_1 G protein beta subunit 2 [Solanum tuber... 203 1e-51
sp|P93563|GBB_SOLTU Guanine nucleotide-binding protein beta subu... 202 3e-51
>gb|AAG12330.1|AF249743_1 heterotrimeric GTP-binding protein subunit beta 1 [Nicotiana
tabacum]
Length = 377
Score = 207 bits (526), Expect = 1e-52
Identities = 97/108 (89%), Positives = 102/108 (93%), Gaps = 1/108 (0%)
Frame = -3
Query: 665 SEDGTCRLFDIRTGHQLQVYYQQHGD-DIPPVTSIAFSISGRLLFAGYANGDCYVWDTLL 489
SEDGTCRLFDIRTGHQLQVYYQ HGD DIP VTS+AFSISGRLLF GY+NGDCYVWDTLL
Sbjct: 267 SEDGTCRLFDIRTGHQLQVYYQPHGDGDIPHVTSMAFSISGRLLFVGYSNGDCYVWDTLL 326
Query: 488 AKVVLNIGSLQDSHEGRISCLGLSADGSALCTGGWDTNLKIWAFGGHR 345
AKVVLN+G +Q+SHEGRISCLGLSADGSALCTG WDTNLKIWAFGGHR
Sbjct: 327 AKVVLNLGGVQNSHEGRISCLGLSADGSALCTGSWDTNLKIWAFGGHR 374
Score = 38.5 bits (88), Expect = 0.082
Identities = 30/103 (29%), Positives = 47/103 (45%), Gaps = 2/103 (1%)
Frame = -3
Query: 665 SEDGTCRLFDIRTGHQLQVYYQQ-HGDDIPPVTSIAFSISG-RLLFAGYANGDCYVWDTL 492
S D TC L+DI TG + V+ + V S++ S S RL +G + +WDT
Sbjct: 176 SGDQTCVLWDITTGLRTSVFGGEFQSGHTADVQSVSISSSNPRLFVSGSCDTTAGLWDT- 234
Query: 491 LAKVVLNIGSLQDSHEGRISCLGLSADGSALCTGGWDTNLKIW 363
+V HEG ++ + S DG+ TG D +++
Sbjct: 235 --RVASRAQRTFYGHEGDVNTVKFSPDGNRFGTGSEDGTCRLF 275
>sp|P93398|GBB2_TOBAC Guanine nucleotide-binding protein beta subunit 2
gi|7441564|pir||T04089 GTP-binding protein beta chain
(clone Gbeta2) - common tobacco
gi|1835163|emb|CAB06619.1| G protein beta subunit
[Nicotiana tabacum]
Length = 377
Score = 206 bits (524), Expect = 2e-52
Identities = 96/108 (88%), Positives = 103/108 (94%), Gaps = 1/108 (0%)
Frame = -3
Query: 665 SEDGTCRLFDIRTGHQLQVYYQQHGD-DIPPVTSIAFSISGRLLFAGYANGDCYVWDTLL 489
S+DGTCRLFDIRTGHQLQVYYQ HGD DIP VTS+AFSISGRLLF GY+NGDCYVWDTLL
Sbjct: 267 SDDGTCRLFDIRTGHQLQVYYQPHGDGDIPHVTSMAFSISGRLLFVGYSNGDCYVWDTLL 326
Query: 488 AKVVLNIGSLQDSHEGRISCLGLSADGSALCTGGWDTNLKIWAFGGHR 345
AKVVLN+G++Q+SHEGRISCLGLSADGSALCTG WDTNLKIWAFGGHR
Sbjct: 327 AKVVLNLGAVQNSHEGRISCLGLSADGSALCTGSWDTNLKIWAFGGHR 374
>sp|P93397|GBB1_TOBAC Guanine nucleotide-binding protein beta subunit 1
gi|7441563|pir||T04086 GTP binding protein beta chain -
common tobacco gi|1835161|emb|CAB06618.1| G protein beta
subunit [Nicotiana tabacum]
Length = 377
Score = 204 bits (518), Expect = 1e-51
Identities = 96/108 (88%), Positives = 101/108 (92%), Gaps = 1/108 (0%)
Frame = -3
Query: 665 SEDGTCRLFDIRTGHQLQVYYQQHGD-DIPPVTSIAFSISGRLLFAGYANGDCYVWDTLL 489
SEDGTCRLFDIRT HQLQVYYQ HGD DIP VTS+AFSISGRLLF GY+NGDCYVWDTLL
Sbjct: 267 SEDGTCRLFDIRTEHQLQVYYQPHGDGDIPHVTSMAFSISGRLLFVGYSNGDCYVWDTLL 326
Query: 488 AKVVLNIGSLQDSHEGRISCLGLSADGSALCTGGWDTNLKIWAFGGHR 345
AKVVLN+G +Q+SHEGRISCLGLSADGSALCTG WDTNLKIWAFGGHR
Sbjct: 327 AKVVLNLGGVQNSHEGRISCLGLSADGSALCTGSWDTNLKIWAFGGHR 374
Score = 36.6 bits (83), Expect = 0.31
Identities = 29/103 (28%), Positives = 46/103 (44%), Gaps = 2/103 (1%)
Frame = -3
Query: 665 SEDGTCRLFDIRTGHQLQVYYQQ-HGDDIPPVTSIAFSISG-RLLFAGYANGDCYVWDTL 492
S D TC L+DI TG + V+ + V S++ S S RL +G + +WDT
Sbjct: 176 SGDQTCVLWDITTGLRTSVFGGEFQSGHTADVQSVSISSSNPRLFVSGSCDTTARLWDT- 234
Query: 491 LAKVVLNIGSLQDSHEGRISCLGLSADGSALCTGGWDTNLKIW 363
+V HEG ++ + DG+ TG D +++
Sbjct: 235 --RVASRAQRTFYGHEGDVNTVKFFPDGNRFGTGSEDGTCRLF 275
>gb|AAL07488.1|AF414114_1 G protein beta subunit 2 [Solanum tuberosum]
Length = 377
Score = 203 bits (517), Expect = 1e-51
Identities = 96/108 (88%), Positives = 102/108 (93%), Gaps = 1/108 (0%)
Frame = -3
Query: 665 SEDGTCRLFDIRTGHQLQVYYQQHGD-DIPPVTSIAFSISGRLLFAGYANGDCYVWDTLL 489
S+DG+CRLFDIRTGHQLQVY Q HGD DIP VTSIAFSISGRLLF GY+NGDCYVWDTLL
Sbjct: 267 SDDGSCRLFDIRTGHQLQVYNQPHGDGDIPHVTSIAFSISGRLLFVGYSNGDCYVWDTLL 326
Query: 488 AKVVLNIGSLQDSHEGRISCLGLSADGSALCTGGWDTNLKIWAFGGHR 345
AKVVLN+GS+Q+SHEGRISCLGLSADGSALCTG WDTNLKIWAFGGHR
Sbjct: 327 AKVVLNLGSVQNSHEGRISCLGLSADGSALCTGSWDTNLKIWAFGGHR 374
>sp|P93563|GBB_SOLTU Guanine nucleotide-binding protein beta subunit
gi|7441557|pir||T07376 G-protein beta chain GB1 - potato
gi|1771734|emb|CAA61106.1| GB1 protein [Solanum
tuberosum]
Length = 377
Score = 202 bits (514), Expect = 3e-51
Identities = 95/108 (87%), Positives = 102/108 (93%), Gaps = 1/108 (0%)
Frame = -3
Query: 665 SEDGTCRLFDIRTGHQLQVYYQQHGD-DIPPVTSIAFSISGRLLFAGYANGDCYVWDTLL 489
S+DG+CRLFDIRTGHQLQVY Q HGD DIP VTS+AFSISGRLLF GY+NGDCYVWDTLL
Sbjct: 267 SDDGSCRLFDIRTGHQLQVYNQPHGDGDIPHVTSMAFSISGRLLFVGYSNGDCYVWDTLL 326
Query: 488 AKVVLNIGSLQDSHEGRISCLGLSADGSALCTGGWDTNLKIWAFGGHR 345
AKVVLN+GS+Q+SHEGRISCLGLSADGSALCTG WDTNLKIWAFGGHR
Sbjct: 327 AKVVLNLGSVQNSHEGRISCLGLSADGSALCTGSWDTNLKIWAFGGHR 374
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 586,386,239
Number of Sequences: 1393205
Number of extensions: 13074663
Number of successful extensions: 38498
Number of sequences better than 10.0: 990
Number of HSP's better than 10.0 without gapping: 34008
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37675
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28855580904
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)