Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
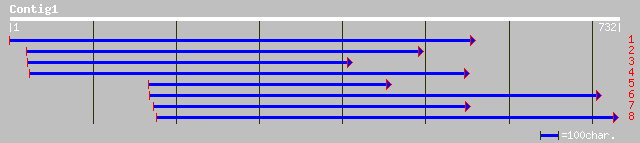
Query= KMC002346A_C01 KMC002346A_c01
(724 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB01310.1| gene_id:MPN9.20~unknown protein [Arabidopsis tha... 72 8e-12
ref|NP_188629.1| unknown protein; protein id: At3g19950.1, suppo... 72 8e-12
dbj|BAC42901.1| unknown protein [Arabidopsis thaliana] 72 8e-12
gb|AAL59019.1|AC087182_2 putative zinc finger protein [Oryza sat... 65 7e-10
ref|NP_700520.1| hypothetical protein [Plasmodium falciparum 3D7... 58 2e-07
>dbj|BAB01310.1| gene_id:MPN9.20~unknown protein [Arabidopsis thaliana]
Length = 386
Score = 72.0 bits (175), Expect = 8e-12
Identities = 45/106 (42%), Positives = 52/106 (48%)
Frame = -2
Query: 723 NSCPVCRHELPTDDPDYEQRAGRRVSTETSGGGGGRSSGDSSQRRGFRVSLPWPFRQSGS 544
NSCPVCR ELPTDDPDYE R S + G G G+ S + Q F + LPWPFR+
Sbjct: 309 NSCPVCRFELPTDDPDYENR-----SQGSQGSGDGQGSVEGQQTPRFSIQLPWPFRR--- 360
Query: 543 SAAETSNVGNANNNSNNNSGNNSGQSNTENRGNQNFESETRQEDLD 406
+ SG+ SG T G N ETR EDLD
Sbjct: 361 ---------------QDGSGSGSGAPGT---GGGNL--ETRGEDLD 386
>ref|NP_188629.1| unknown protein; protein id: At3g19950.1, supported by cDNA:
gi_20465262 [Arabidopsis thaliana]
gi|20465263|gb|AAM19951.1| AT3g19950/MPN9_19
[Arabidopsis thaliana] gi|23308365|gb|AAN18152.1|
At3g19950/MPN9_19 [Arabidopsis thaliana]
Length = 328
Score = 72.0 bits (175), Expect = 8e-12
Identities = 45/106 (42%), Positives = 52/106 (48%)
Frame = -2
Query: 723 NSCPVCRHELPTDDPDYEQRAGRRVSTETSGGGGGRSSGDSSQRRGFRVSLPWPFRQSGS 544
NSCPVCR ELPTDDPDYE R S + G G G+ S + Q F + LPWPFR+
Sbjct: 251 NSCPVCRFELPTDDPDYENR-----SQGSQGSGDGQGSVEGQQTPRFSIQLPWPFRR--- 302
Query: 543 SAAETSNVGNANNNSNNNSGNNSGQSNTENRGNQNFESETRQEDLD 406
+ SG+ SG T G N ETR EDLD
Sbjct: 303 ---------------QDGSGSGSGAPGT---GGGNL--ETRGEDLD 328
>dbj|BAC42901.1| unknown protein [Arabidopsis thaliana]
Length = 121
Score = 72.0 bits (175), Expect = 8e-12
Identities = 45/106 (42%), Positives = 52/106 (48%)
Frame = -2
Query: 723 NSCPVCRHELPTDDPDYEQRAGRRVSTETSGGGGGRSSGDSSQRRGFRVSLPWPFRQSGS 544
NSCPVCR ELPTDDPDYE R S + G G G+ S + Q F + LPWPFR+
Sbjct: 44 NSCPVCRFELPTDDPDYENR-----SQGSQGSGDGQGSVEGQQTPRFSIQLPWPFRR--- 95
Query: 543 SAAETSNVGNANNNSNNNSGNNSGQSNTENRGNQNFESETRQEDLD 406
+ SG+ SG T G N ETR EDLD
Sbjct: 96 ---------------QDGSGSGSGAPGT---GGGNL--ETRGEDLD 121
>gb|AAL59019.1|AC087182_2 putative zinc finger protein [Oryza sativa]
Length = 370
Score = 65.5 bits (158), Expect = 7e-10
Identities = 40/107 (37%), Positives = 54/107 (50%), Gaps = 16/107 (14%)
Frame = -2
Query: 723 NSCPVCRHELPTDDPDYEQRAGRRVST--ETSGGGGGRSSGDSSQ--------------R 592
NSCP+CR ELPTDDPDYE GR+ S T+G G +SG S+
Sbjct: 271 NSCPICRFELPTDDPDYE---GRKKSNPQPTAGVDAGAASGSSTAAEEREESGESARLVE 327
Query: 591 RGFRVSLPWPFRQSGSSAAETSNVGNANNNSNNNSGNNSGQSNTENR 451
R F VSLPWPF GS + + +N SG+ G ++++ +
Sbjct: 328 RRFNVSLPWPFSGLGSQTPQQ----DGSNGGAGASGSKDGGASSDKK 370
>ref|NP_700520.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23494909|gb|AAN35244.1|AE014829_44 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 1130
Score = 57.8 bits (138), Expect = 2e-07
Identities = 32/107 (29%), Positives = 47/107 (43%), Gaps = 3/107 (2%)
Frame = -2
Query: 723 NSCPVCRHELPTDDPDYE---QRAGRRVSTETSGGGGGRSSGDSSQRRGFRVSLPWPFRQ 553
NSCP CR ELPTDD +Y + R+++E S + + +
Sbjct: 400 NSCPTCRFELPTDDQEYNCKREELRERLNSEMSRNNTYNGTDNDNN-------------- 445
Query: 552 SGSSAAETSNVGNANNNSNNNSGNNSGQSNTENRGNQNFESETRQED 412
+ ++ N N NNN NNN+ NN+ +N N N N +S D
Sbjct: 446 NNNNDNNNDNNNNDNNNDNNNNDNNNDNNNDNNNNNSNNDSNNNNND 492
Score = 42.4 bits (98), Expect = 0.007
Identities = 26/101 (25%), Positives = 43/101 (41%), Gaps = 9/101 (8%)
Frame = -2
Query: 714 PVCRHELPTDDPDYEQRAGRRVSTETSGGGGGRSSGDSSQRR---------GFRVSLPWP 562
P+ H L T E R+G + + G +S +++ R G+R ++P
Sbjct: 50 PLSFHSLHTSRDVGESRSGPNSRNDNNNNDGNNNSNNTNSRHYNYRNMNNFGYRTNIP-- 107
Query: 561 FRQSGSSAAETSNVGNANNNSNNNSGNNSGQSNTENRGNQN 439
S N NNN+NN+ G+N+ + N+ N N N
Sbjct: 108 ----------RSRRNNHNNNNNNDDGSNNNRHNSSNSNNNN 138
Score = 41.2 bits (95), Expect = 0.015
Identities = 24/78 (30%), Positives = 38/78 (47%), Gaps = 2/78 (2%)
Frame = -2
Query: 639 TSGGGGGRSSGDSSQRRGFR--VSLPWPFRQSGSSAAETSNVGNANNNSNNNSGNNSGQS 466
T+ G +S D ++ R + P + ++ +N N NNN+NN+ GN +
Sbjct: 182 TAAVGSSNNSNDINRHNNNRNNTTTPTTNNNNNNNNNNNNNNNNNNNNNNNHMGNRNSSR 241
Query: 465 NTENRGNQNFESETRQED 412
N EN N+ E+ET Q D
Sbjct: 242 NNENT-NETNENETSQND 258
Score = 35.0 bits (79), Expect = 1.1
Identities = 14/37 (37%), Positives = 21/37 (55%)
Frame = -2
Query: 549 GSSAAETSNVGNANNNSNNNSGNNSGQSNTENRGNQN 439
G +T NV N NNN N+N N++ ++ +N N N
Sbjct: 763 GQRNNDTDNVRNNNNNDNDNKNNDNKNNDNDNNNNNN 799
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 625,268,209
Number of Sequences: 1393205
Number of extensions: 14403588
Number of successful extensions: 237841
Number of sequences better than 10.0: 2411
Number of HSP's better than 10.0 without gapping: 71518
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 142984
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33780557640
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)