Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002345A_C01 KMC002345A_c01
(571 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
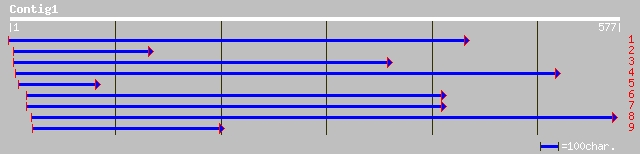
gb|AAF81279.1|AF257178_1 C-14 sterol reductase [Arabidopsis thal... 220 8e-57
ref|NP_566975.1| nuclear envelope membrane protein - like; prote... 220 8e-57
pir||T47551 nuclear envelope membrane protein-like - Arabidopsis... 220 8e-57
gb|AAG30271.1|AF308470_1 sterol C-14 reductase [Dictyostelium di... 129 2e-29
dbj|BAB29187.1| unnamed protein product [Mus musculus] 128 6e-29
>gb|AAF81279.1|AF257178_1 C-14 sterol reductase [Arabidopsis thaliana]
gi|9714052|emb|CAC01296.1| sterol C-14 reductase
(FACKEL) [Arabidopsis thaliana]
Length = 365
Score = 220 bits (561), Expect = 8e-57
Identities = 99/113 (87%), Positives = 104/113 (91%)
Frame = -2
Query: 570 VFRGADKHKHEFK*NPKAPIWVKPPIVIGGKLLASGYWGVARHCNYLGDLLLALSFSLPC 391
VFRGA+K KH FK NPK PIW KPP+V+GGKLL SGYWG+ARHCNYLGDL+LALSFSLPC
Sbjct: 253 VFRGANKQKHIFKKNPKTPIWGKPPVVVGGKLLVSGYWGIARHCNYLGDLMLALSFSLPC 312
Query: 390 GISSPVPYFYPIYLLILLIWRERRDEARCAEKYKEIWAEYRRTVPWRILPYVY 232
GISSPVPYFYPIYLLILLIWRERRDE RCAEKYKEIWAEY R VPWRILPYVY
Sbjct: 313 GISSPVPYFYPIYLLILLIWRERRDEVRCAEKYKEIWAEYLRLVPWRILPYVY 365
>ref|NP_566975.1| nuclear envelope membrane protein - like; protein id: At3g52940.1,
supported by cDNA: gi_17473515, supported by cDNA:
gi_8980703 [Arabidopsis thaliana]
gi|20138022|sp|Q9LDR4|ER24_ARATH Delta(14)-sterol
reductase (C-14 sterol reductase) (Sterol C14-reductase)
(FACKEL protein) gi|8980704|gb|AAF82282.1| sterol C-14
reductase FACKEL [Arabidopsis thaliana]
gi|8980706|gb|AAF82283.1|AF256536_1 sterol C-14
reductase FACKEL [Arabidopsis thaliana]
gi|9082182|gb|AAF82768.1| C-14 sterol reductase
[Arabidopsis thaliana] gi|17473516|gb|AAL38381.1|
AT3g52940/F8J2_111 [Arabidopsis thaliana]
gi|22137270|gb|AAM91480.1| AT3g52940/F8J2_111
[Arabidopsis thaliana]
Length = 369
Score = 220 bits (561), Expect = 8e-57
Identities = 99/113 (87%), Positives = 104/113 (91%)
Frame = -2
Query: 570 VFRGADKHKHEFK*NPKAPIWVKPPIVIGGKLLASGYWGVARHCNYLGDLLLALSFSLPC 391
VFRGA+K KH FK NPK PIW KPP+V+GGKLL SGYWG+ARHCNYLGDL+LALSFSLPC
Sbjct: 257 VFRGANKQKHIFKKNPKTPIWGKPPVVVGGKLLVSGYWGIARHCNYLGDLMLALSFSLPC 316
Query: 390 GISSPVPYFYPIYLLILLIWRERRDEARCAEKYKEIWAEYRRTVPWRILPYVY 232
GISSPVPYFYPIYLLILLIWRERRDE RCAEKYKEIWAEY R VPWRILPYVY
Sbjct: 317 GISSPVPYFYPIYLLILLIWRERRDEVRCAEKYKEIWAEYLRLVPWRILPYVY 369
>pir||T47551 nuclear envelope membrane protein-like - Arabidopsis thaliana
Length = 317
Score = 220 bits (561), Expect = 8e-57
Identities = 99/113 (87%), Positives = 104/113 (91%)
Frame = -2
Query: 570 VFRGADKHKHEFK*NPKAPIWVKPPIVIGGKLLASGYWGVARHCNYLGDLLLALSFSLPC 391
VFRGA+K KH FK NPK PIW KPP+V+GGKLL SGYWG+ARHCNYLGDL+LALSFSLPC
Sbjct: 205 VFRGANKQKHIFKKNPKTPIWGKPPVVVGGKLLVSGYWGIARHCNYLGDLMLALSFSLPC 264
Query: 390 GISSPVPYFYPIYLLILLIWRERRDEARCAEKYKEIWAEYRRTVPWRILPYVY 232
GISSPVPYFYPIYLLILLIWRERRDE RCAEKYKEIWAEY R VPWRILPYVY
Sbjct: 265 GISSPVPYFYPIYLLILLIWRERRDEVRCAEKYKEIWAEYLRLVPWRILPYVY 317
>gb|AAG30271.1|AF308470_1 sterol C-14 reductase [Dictyostelium discoideum]
gi|11096271|gb|AAG30272.1| putative sterol C-14
reductase [Dictyostelium discoideum]
Length = 292
Score = 129 bits (325), Expect = 2e-29
Identities = 58/110 (52%), Positives = 78/110 (70%)
Frame = -2
Query: 570 VFRGADKHKHEFK*NPKAPIWVKPPIVIGGKLLASGYWGVARHCNYLGDLLLALSFSLPC 391
VFR ++ KH+FK NPK+ IW K P I GKLL SG+WG+ R NYLGD ++A SFS PC
Sbjct: 180 VFRISNSQKHQFKTNPKSLIWGKKPETIAGKLLVSGFWGILRKPNYLGDWIVAFSFSFPC 239
Query: 390 GISSPVPYFYPIYLLILLIWRERRDEARCAEKYKEIWAEYRRTVPWRILP 241
++P+ Y YPIYL++L R+ RD +C++KY E+W EY R VP+ +LP
Sbjct: 240 LFNTPIVYLYPIYLVVLNSHRQWRDHQKCSKKYGELWKEYCRRVPYVVLP 289
>dbj|BAB29187.1| unnamed protein product [Mus musculus]
Length = 358
Score = 128 bits (321), Expect = 6e-29
Identities = 59/116 (50%), Positives = 79/116 (67%), Gaps = 3/116 (2%)
Frame = -2
Query: 570 VFRGADKHKHEFK*NPKAPIWV---KPPIVIGGKLLASGYWGVARHCNYLGDLLLALSFS 400
+FRGA+ K+ F+ NP P P G +LL SG+WG+ RH NYLGDL++AL++S
Sbjct: 243 IFRGANSQKNTFRKNPSDPSVAGLETIPTATGRQLLVSGWWGMVRHPNYLGDLIMALAWS 302
Query: 399 LPCGISSPVPYFYPIYLLILLIWRERRDEARCAEKYKEIWAEYRRTVPWRILPYVY 232
LPCG+S +PYFY +Y LL+ RE RDE +C +KY W EY + VP+RI+PYVY
Sbjct: 303 LPCGLSHLLPYFYVLYFTALLVHREARDEQQCLQKYGRAWQEYCKRVPYRIIPYVY 358
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 476,426,403
Number of Sequences: 1393205
Number of extensions: 10181675
Number of successful extensions: 30703
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 29334
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30588
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)