Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002330A_C01 KMC002330A_c01
(714 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
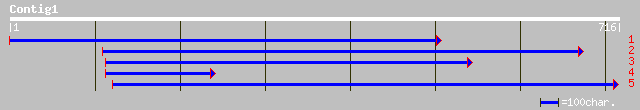
ref|NP_191190.2| glycosyl hydrolase family 27 (alpha-galactosida... 194 2e-65
pir||T47748 alpha-galactosidase-like protein - Arabidopsis thali... 194 2e-65
sp|Q42656|AGAL_COFAR ALPHA-GALACTOSIDASE PRECURSOR (MELIBIASE) (... 157 2e-44
dbj|BAC66445.1| alpha-galactosidase [Helianthus annuus] 157 5e-43
pir||T06388 alpha-galactosidase (EC 3.2.1.22) - soybean gi|92757... 152 5e-43
>ref|NP_191190.2| glycosyl hydrolase family 27 (alpha-galactosidase/melibiase);
protein id: At3g56310.1, supported by cDNA: gi_18377734,
supported by cDNA: gi_21281027 [Arabidopsis thaliana]
gi|18377735|gb|AAL67017.1| putative alpha-galactosidase
[Arabidopsis thaliana] gi|21281028|gb|AAM45068.1|
putative alpha-galactosidase [Arabidopsis thaliana]
Length = 437
Score = 194 bits (493), Expect(2) = 2e-65
Identities = 88/115 (76%), Positives = 101/115 (87%)
Frame = -3
Query: 712 GWNDPDMLEVGNGGMTYQEYRAHFSIWALAKAPLLIGCDIRNMTAETLEIISNKEVIAIN 533
GWNDPDMLE+GNGGMTY+EYR HFSIWAL KAPLLIGCD+RNMTAETLEI+SNKE+IA+N
Sbjct: 274 GWNDPDMLEIGNGGMTYEEYRGHFSIWALMKAPLLIGCDVRNMTAETLEILSNKEIIAVN 333
Query: 532 QDSLGVQGRKVQVAGKDGCSQVWAGPLSGDRWVVALWNRCSKVANITSFLGSTWD 368
QD LGVQGRK+Q G++ C QVW+GPLSGDR VVALWNRCS+ A IT ++WD
Sbjct: 334 QDPLGVQGRKIQANGENDCQQVWSGPLSGDRMVVALWNRCSEPATIT----ASWD 384
Score = 77.4 bits (189), Expect(2) = 2e-65
Identities = 34/55 (61%), Positives = 42/55 (75%)
Frame = -1
Query: 390 ASWEALGIESGVYVTVRDLWQHKVVTEDAVSSFSAQVDTHDSHLYIFTPSTVSLS 226
ASW+ +G+ES + V+VRDLWQHK VTE+ SF AQVD HD H+Y+ TP TVS S
Sbjct: 381 ASWDMIGLESTISVSVRDLWQHKDVTENTSGSFEAQVDAHDCHMYVLTPQTVSHS 435
>pir||T47748 alpha-galactosidase-like protein - Arabidopsis thaliana
gi|7572929|emb|CAB87430.1| alpha-galactosidase-like
protein [Arabidopsis thaliana]
Length = 434
Score = 194 bits (493), Expect(2) = 2e-65
Identities = 88/115 (76%), Positives = 101/115 (87%)
Frame = -3
Query: 712 GWNDPDMLEVGNGGMTYQEYRAHFSIWALAKAPLLIGCDIRNMTAETLEIISNKEVIAIN 533
GWNDPDMLE+GNGGMTY+EYR HFSIWAL KAPLLIGCD+RNMTAETLEI+SNKE+IA+N
Sbjct: 271 GWNDPDMLEIGNGGMTYEEYRGHFSIWALMKAPLLIGCDVRNMTAETLEILSNKEIIAVN 330
Query: 532 QDSLGVQGRKVQVAGKDGCSQVWAGPLSGDRWVVALWNRCSKVANITSFLGSTWD 368
QD LGVQGRK+Q G++ C QVW+GPLSGDR VVALWNRCS+ A IT ++WD
Sbjct: 331 QDPLGVQGRKIQANGENDCQQVWSGPLSGDRMVVALWNRCSEPATIT----ASWD 381
Score = 77.4 bits (189), Expect(2) = 2e-65
Identities = 34/55 (61%), Positives = 42/55 (75%)
Frame = -1
Query: 390 ASWEALGIESGVYVTVRDLWQHKVVTEDAVSSFSAQVDTHDSHLYIFTPSTVSLS 226
ASW+ +G+ES + V+VRDLWQHK VTE+ SF AQVD HD H+Y+ TP TVS S
Sbjct: 378 ASWDMIGLESTISVSVRDLWQHKDVTENTSGSFEAQVDAHDCHMYVLTPQTVSHS 432
>sp|Q42656|AGAL_COFAR ALPHA-GALACTOSIDASE PRECURSOR (MELIBIASE) (ALPHA-D-GALACTOSIDE
GALACTOHYDROLASE) gi|11264279|pir||T50781
alpha-galactosidase (EC 3.2.1.22) [imported] - coffee
gi|504489|gb|AAA33022.1| alpha-galactosidase
Length = 378
Score = 157 bits (396), Expect(2) = 2e-44
Identities = 76/109 (69%), Positives = 86/109 (78%)
Frame = -3
Query: 712 GWNDPDMLEVGNGGMTYQEYRAHFSIWALAKAPLLIGCDIRNMTAETLEIISNKEVIAIN 533
GWNDPDMLEVGNGGMT EYR+HFSIWALAKAPLLIGCDIR+M T +++SN EVIA+N
Sbjct: 226 GWNDPDMLEVGNGGMTTTEYRSHFSIWALAKAPLLIGCDIRSMDGATFQLLSNAEVIAVN 285
Query: 532 QDSLGVQGRKVQVAGKDGCSQVWAGPLSGDRWVVALWNRCSKVANITSF 386
QD LGVQG KV+ G +VWAGPLSG R VALWNR S A IT++
Sbjct: 286 QDKLGVQGNKVKTYGD---LEVWAGPLSGKRVAVALWNRGSSTATITAY 331
Score = 44.7 bits (104), Expect(2) = 2e-44
Identities = 23/50 (46%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Frame = -1
Query: 390 ASWEALGIESGVYVTVRDLWQHKVVTEDAVSS-FSAQVDTHDSHLYIFTP 244
A W +G+ S V RDLW H TE +V SA VD HDS +Y+ TP
Sbjct: 330 AYWSDVGLPSTAVVNARDLWAHS--TEKSVKGQISAAVDAHDSKMYVLTP 377
>dbj|BAC66445.1| alpha-galactosidase [Helianthus annuus]
Length = 428
Score = 157 bits (396), Expect(2) = 5e-43
Identities = 74/109 (67%), Positives = 93/109 (84%)
Frame = -3
Query: 712 GWNDPDMLEVGNGGMTYQEYRAHFSIWALAKAPLLIGCDIRNMTAETLEIISNKEVIAIN 533
GWNDPDMLEVGNGGMT +EYR+HFSIWALAKAPLL+GCD+R+M+ ET +I+SN+EVIA+N
Sbjct: 276 GWNDPDMLEVGNGGMTTEEYRSHFSIWALAKAPLLVGCDMRSMSKETHDILSNREVIAVN 335
Query: 532 QDSLGVQGRKVQVAGKDGCSQVWAGPLSGDRWVVALWNRCSKVANITSF 386
QDSLGVQG+KV+ K+G +VWAGPL+ ++ V LWNR S A IT++
Sbjct: 336 QDSLGVQGKKVK---KNGDLEVWAGPLAHNKVAVILWNRGSSRAQITAY 381
Score = 40.0 bits (92), Expect(2) = 5e-43
Identities = 18/49 (36%), Positives = 25/49 (50%)
Frame = -1
Query: 390 ASWEALGIESGVYVTVRDLWQHKVVTEDAVSSFSAQVDTHDSHLYIFTP 244
A W +G+ S V VRDLW H+ SA V++H +Y+ TP
Sbjct: 380 AYWSDIGLNSTTVVNVRDLWAHR-TQRSVKGQISATVESHACKMYVLTP 427
>pir||T06388 alpha-galactosidase (EC 3.2.1.22) - soybean
gi|927575|gb|AAA73963.1| alpha galactosidase
Length = 422
Score = 152 bits (384), Expect(2) = 5e-43
Identities = 73/108 (67%), Positives = 86/108 (79%)
Frame = -3
Query: 712 GWNDPDMLEVGNGGMTYQEYRAHFSIWALAKAPLLIGCDIRNMTAETLEIISNKEVIAIN 533
GWNDPDMLEVGNGGMT +EYRAHFSIW+LAKAPLLIGCDIR + A T E++SNKEVIA+N
Sbjct: 270 GWNDPDMLEVGNGGMTTEEYRAHFSIWSLAKAPLLIGCDIRALDATTKELLSNKEVIAVN 329
Query: 532 QDSLGVQGRKVQVAGKDGCSQVWAGPLSGDRWVVALWNRCSKVANITS 389
QD LGVQG+KV+ +VWAGPLS ++ V LWNR S A +T+
Sbjct: 330 QDKLGVQGKKVKSTND---LEVWAGPLSNNKVAVILWNRSSSKAKVTA 374
Score = 44.7 bits (104), Expect(2) = 5e-43
Identities = 21/51 (41%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Frame = -1
Query: 390 ASWEALGIESGVYVTVRDLWQHKVVTEDAVS-SFSAQVDTHDSHLYIFTPS 241
ASW +G++ G V RDLW H T+ +VS SA++D+H +Y+ TP+
Sbjct: 374 ASWSDIGLKPGTSVEARDLWAHS--TQSSVSGEISAELDSHACKMYVVTPN 422
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 613,333,878
Number of Sequences: 1393205
Number of extensions: 13146218
Number of successful extensions: 30355
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 29347
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30295
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32936043699
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)