Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002329A_C01 KMC002329A_c01
(624 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
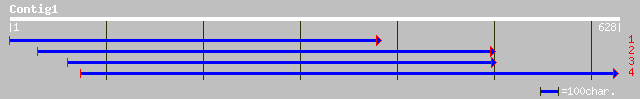
ref|NP_172447.1| putative U2 small nuclear ribonucleoprotein A (... 74 2e-12
pir||S30580 U2 snRNP protein A' - Arabidopsis thaliana gi|17669|... 74 2e-12
gb|AAH22816.1| Unknown (protein for MGC:39248) [Homo sapiens] 39 0.042
ref|NP_003081.1| u2 small nuclear ribonucleoprotein polypeptide ... 39 0.042
emb|CAA06160.1| U2 snRNP-specific A' protein [Salmo salar] 39 0.072
>ref|NP_172447.1| putative U2 small nuclear ribonucleoprotein A (U2 SNRNP-A); protein
id: At1g09760.1, supported by cDNA: gi_16649064
[Arabidopsis thaliana] gi|19884091|sp|P43333|RU2A_ARATH
U2 small nuclear ribonucleoprotein A' (U2 snRNP-A')
gi|25402575|pir||E86231 hypothetical protein [imported]
- Arabidopsis thaliana gi|2160183|gb|AAB60746.1|
Identical to A. thaliana U2 SnRNP-specific A' protein
(gb|X69137). ESTs gb|ATTS0705, gb|ATTS0339 come from
this gene. [Arabidopsis thaliana]
gi|16649065|gb|AAL24384.1| SnRNP-specific A protein
[Arabidopsis thaliana] gi|23197870|gb|AAN15462.1|
SnRNP-specific A protein [Arabidopsis thaliana]
Length = 249
Score = 73.9 bits (180), Expect = 2e-12
Identities = 37/57 (64%), Positives = 45/57 (78%)
Frame = -1
Query: 618 ETPNVSEATEDXQTPXVVAPTPEQIIAIKAAIVNSQTLEEVARLEKALKSGLLPEDL 448
E VSE E+ +TP VVAPT EQI+AIKAAI+NSQT+EE+ARLE+ALK G +P L
Sbjct: 178 EVKKVSETAENPETPKVVAPTAEQILAIKAAIINSQTIEEIARLEQALKFGQVPAGL 234
>pir||S30580 U2 snRNP protein A' - Arabidopsis thaliana gi|17669|emb|CAA48890.1|
U2 small nuclear ribonucleoprotein A' [Arabidopsis
thaliana]
Length = 249
Score = 73.9 bits (180), Expect = 2e-12
Identities = 37/57 (64%), Positives = 45/57 (78%)
Frame = -1
Query: 618 ETPNVSEATEDXQTPXVVAPTPEQIIAIKAAIVNSQTLEEVARLEKALKSGLLPEDL 448
E VSE E+ +TP VVAPT EQI+AIKAAI+NSQT+EE+ARLE+ALK G +P L
Sbjct: 178 EVKKVSETAENPETPKVVAPTAEQILAIKAAIINSQTIEEIARLEQALKFGQVPAGL 234
>gb|AAH22816.1| Unknown (protein for MGC:39248) [Homo sapiens]
Length = 255
Score = 39.3 bits (90), Expect = 0.042
Identities = 25/62 (40%), Positives = 33/62 (52%)
Frame = -1
Query: 561 PTPEQIIAIKAAIVNSQTLEEVARLEKALKSGLLPEDLKSLNGNVTLDNVNEKGEDVVHD 382
P+P + AIK AI N+ TL EV RL+ L+SG +P + T D E ED V +
Sbjct: 196 PSPGDVEAIKNAIANASTLAEVERLKGLLQSGQIPG--RERRSGPTDDGEEEMEEDTVTN 253
Query: 381 ES 376
S
Sbjct: 254 GS 255
>ref|NP_003081.1| u2 small nuclear ribonucleoprotein polypeptide A' [Homo sapiens]
gi|134094|sp|P09661|RU2A_HUMAN U2 small nuclear
ribonucleoprotein A' (U2 snRNP-A') gi|88963|pir||S03616
U2 snRNP protein A' - human gi|37547|emb|CAA31838.1| U2
snRNP-specific A' protein (AA 1-255) [Homo sapiens]
Length = 255
Score = 39.3 bits (90), Expect = 0.042
Identities = 25/62 (40%), Positives = 33/62 (52%)
Frame = -1
Query: 561 PTPEQIIAIKAAIVNSQTLEEVARLEKALKSGLLPEDLKSLNGNVTLDNVNEKGEDVVHD 382
P+P + AIK AI N+ TL EV RL+ L+SG +P + T D E ED V +
Sbjct: 196 PSPGDVEAIKNAIANASTLAEVERLKGLLQSGQIPG--RERRSGPTDDGEEEMEEDTVTN 253
Query: 381 ES 376
S
Sbjct: 254 GS 255
>emb|CAA06160.1| U2 snRNP-specific A' protein [Salmo salar]
Length = 339
Score = 38.5 bits (88), Expect = 0.072
Identities = 23/73 (31%), Positives = 39/73 (52%), Gaps = 1/73 (1%)
Frame = -1
Query: 567 VAPTPEQIIAIKAAIVNSQTLEEVARLEKALKSGLLP-EDLKSLNGNVTLDNVNEKGEDV 391
+ P+P + AIK AI N+ +L EV RL+ L++G +P +++ + + E+ V
Sbjct: 195 MGPSPADVEAIKNAIANASSLAEVERLKGMLQAGQIPGREVRQVPPEMVEVEEEEEENGV 254
Query: 390 VHDESNDTREQRN 352
VH E E+ N
Sbjct: 255 VHMEREIQTEEGN 267
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,986,220
Number of Sequences: 1393205
Number of extensions: 10058989
Number of successful extensions: 31404
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 29547
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31265
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)