Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002321A_C01 KMC002321A_c01
(598 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
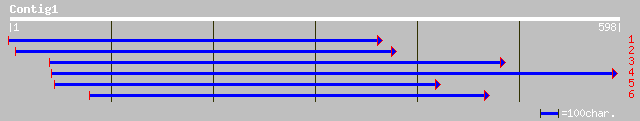
ref|NP_172643.1| alkylation repair-like protein; protein id: At1... 214 5e-55
sp|Q9SA98|ALKB_ARATH Alkylated DNA repair protein alkB homolog g... 207 1e-52
gb|EAA06442.1| agCP13616 [Anopheles gambiae str. PEST] 113 2e-24
gb|AAH25787.1| alkylation repair; alkB homolog [Homo sapiens] 100 2e-20
sp|Q13686|ALKB_HUMAN Alkylated DNA repair protein alkB homolog g... 100 2e-20
>ref|NP_172643.1| alkylation repair-like protein; protein id: At1g11780.1
[Arabidopsis thaliana]
Length = 345
Score = 214 bits (546), Expect = 5e-55
Identities = 106/140 (75%), Positives = 119/140 (84%)
Frame = -1
Query: 598 LCELAKQLAKPALPAGVEFRPEAAIVNYFTSGDTLGGHLDDMEADWSKPIVSLSLGCKAI 419
LC+LAK A A+P G EFRPE AIVNYF GDTLGGHLDDMEADWSKPIVS+SLGCKAI
Sbjct: 206 LCQLAKTHAAIAMPDGEEFRPEGAIVNYFGIGDTLGGHLDDMEADWSKPIVSMSLGCKAI 265
Query: 418 FLLGGKSKEDLPLAMFLRSGDVVLMAGEARECFHGVPRIFTDKENSEIGHLETLLAHQDD 239
FLLGGKSK+D P AM+LRSGDVVLMAGEARECFHG+PRIFT +EN++IG LE+ L+H+
Sbjct: 266 FLLGGKSKDDPPHAMYLRSGDVVLMAGEARECFHGIPRIFTGEENADIGALESELSHESG 325
Query: 238 LCILNYIQTSRININIRQVF 179
YI+TSRININIRQVF
Sbjct: 326 HFFAEYIKTSRININIRQVF 345
>sp|Q9SA98|ALKB_ARATH Alkylated DNA repair protein alkB homolog gi|25402684|pir||G86251
protein F25C20.6 [imported] - Arabidopsis thaliana
gi|4835778|gb|AAD30244.1|AC007296_5 F25C20.6
[Arabidopsis thaliana]
Length = 354
Score = 207 bits (526), Expect = 1e-52
Identities = 106/149 (71%), Positives = 119/149 (79%), Gaps = 9/149 (6%)
Frame = -1
Query: 598 LCELAKQLAKPALPAGVEFRPEAAIVNYFTSGDTLGGHLDDMEADWSKPIVSLSLGCKAI 419
LC+LAK A A+P G EFRPE AIVNYF GDTLGGHLDDMEADWSKPIVS+SLGCKAI
Sbjct: 206 LCQLAKTHAAIAMPDGEEFRPEGAIVNYFGIGDTLGGHLDDMEADWSKPIVSMSLGCKAI 265
Query: 418 FLLGGKSKEDLPLAMFLRSGDVVLMAGEARECFH---------GVPRIFTDKENSEIGHL 266
FLLGGKSK+D P AM+LRSGDVVLMAGEARECFH G+PRIFT +EN++IG L
Sbjct: 266 FLLGGKSKDDPPHAMYLRSGDVVLMAGEARECFHGNLLHFQLDGIPRIFTGEENADIGAL 325
Query: 265 ETLLAHQDDLCILNYIQTSRININIRQVF 179
E+ L+H+ YI+TSRININIRQVF
Sbjct: 326 ESELSHESGHFFAEYIKTSRININIRQVF 354
>gb|EAA06442.1| agCP13616 [Anopheles gambiae str. PEST]
Length = 286
Score = 113 bits (283), Expect = 2e-24
Identities = 66/149 (44%), Positives = 86/149 (57%), Gaps = 11/149 (7%)
Frame = -1
Query: 595 CELAKQLAKPALPAGVE-FRPEAAIVNYFTSGDTLGGHLDDMEADWSKPIVSLSLGCKAI 419
CEL + A G + F PEAAIVNY+ +G TL GH D E D + P+ S S G A+
Sbjct: 124 CELGALVRYVATTLGYDRFSPEAAIVNYYPAGATLAGHTDHSEDDQTAPLFSFSFGQPAV 183
Query: 418 FLLGGKSKEDLPLAMFLRSGDVVLMAGEARECFHGVPRIFTDKENSE----------IGH 269
FL+GG S+E+ P A+ LRSGD+V+M G +R C+H VPR+ D E E +
Sbjct: 184 FLIGGTSREEHPDALLLRSGDIVVMTGASRLCYHAVPRVCIDAELPEGLGCSAARWAVLD 243
Query: 268 LETLLAHQDDLCILNYIQTSRININIRQV 182
E A Q + Y+Q SRININ+RQV
Sbjct: 244 AERPGAVQWGAAVEEYMQHSRININVRQV 272
>gb|AAH25787.1| alkylation repair; alkB homolog [Homo sapiens]
Length = 389
Score = 99.8 bits (247), Expect = 2e-20
Identities = 54/136 (39%), Positives = 81/136 (58%), Gaps = 14/136 (10%)
Frame = -1
Query: 547 EFRPEAAIVNYFTSGDTLGGHLDDMEADWSKPIVSLSLGCKAIFLLGGKSKEDLPLAMFL 368
+FR EA I+NY+ TLG H+D E D SKP++S S G AIFLLGG +++ P AMF+
Sbjct: 211 DFRAEAGILNYYRLDSTLGIHVDRSELDHSKPLLSFSFGQSAIFLLGGLQRDEAPTAMFM 270
Query: 367 RSGDVVLMAGEARECFHGVPRIFTDKENSEIGH-LETLL-------------AHQDDLCI 230
SGD+++M+G +R H VPR+ + E + H LE L + +D
Sbjct: 271 HSGDIMIMSGFSRLLNHAVPRVLPNPEGEGLPHCLEAPLPAVLPRDSMVEPCSMEDWQVC 330
Query: 229 LNYIQTSRININIRQV 182
+Y++T+R+N+ +RQV
Sbjct: 331 ASYLKTARVNMTVRQV 346
>sp|Q13686|ALKB_HUMAN Alkylated DNA repair protein alkB homolog
gi|6015189|gb|AAF01478.1|AC008044_1 ABH [Homo sapiens]
Length = 389
Score = 99.8 bits (247), Expect = 2e-20
Identities = 54/136 (39%), Positives = 81/136 (58%), Gaps = 14/136 (10%)
Frame = -1
Query: 547 EFRPEAAIVNYFTSGDTLGGHLDDMEADWSKPIVSLSLGCKAIFLLGGKSKEDLPLAMFL 368
+FR EA I+NY+ TLG H+D E D SKP++S S G AIFLLGG +++ P AMF+
Sbjct: 211 DFRAEAGILNYYRLDSTLGIHVDRSELDHSKPLLSFSFGQSAIFLLGGLQRDEAPTAMFM 270
Query: 367 RSGDVVLMAGEARECFHGVPRIFTDKENSEIGH-LETLL-------------AHQDDLCI 230
SGD+++M+G +R H VPR+ + E + H LE L + +D
Sbjct: 271 HSGDIMIMSGFSRLLNHAVPRVLPNPEGEGLPHCLEAPLPAVLPRDSMVEPCSMEDWQVC 330
Query: 229 LNYIQTSRININIRQV 182
+Y++T+R+N+ +RQV
Sbjct: 331 ASYLKTARVNMTVRQV 346
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 475,935,253
Number of Sequences: 1393205
Number of extensions: 10162449
Number of successful extensions: 23383
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 22554
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23332
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)