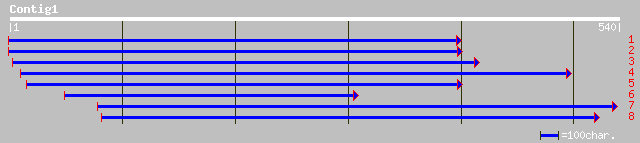
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002314A_C01 KMC002314A_c01
(538 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_193515.1| hypothetical protein; protein id: At4g17800.1, ... 80 2e-14
ref|NP_181070.1| putative AT-hook DNA-binding protein; protein i... 69 4e-11
ref|NP_566232.1| expressed protein; protein id: At3g04570.1, sup... 52 5e-06
emb|CAC42230.1| Krueppel-like protein [Plasmodium falciparum 3D7] 48 9e-05
gb|EAA13112.1| ebiP1409 [Anopheles gambiae str. PEST] 48 9e-05
>ref|NP_193515.1| hypothetical protein; protein id: At4g17800.1, supported by cDNA:
gi_17933298, supported by cDNA: gi_20453386 [Arabidopsis
thaliana] gi|7485086|pir||C71448 hypothetical protein -
Arabidopsis thaliana gi|2245139|emb|CAB10560.1|
hypothetical protein [Arabidopsis thaliana]
gi|7268533|emb|CAB78783.1| hypothetical protein
[Arabidopsis thaliana]
gi|17933299|gb|AAL48232.1|AF446359_1 AT4g17800/dl4935c
[Arabidopsis thaliana] gi|20453387|gb|AAM19932.1|
AT4g17800/dl4935c [Arabidopsis thaliana]
Length = 292
Score = 80.1 bits (196), Expect = 2e-14
Identities = 46/79 (58%), Positives = 51/79 (64%), Gaps = 7/79 (8%)
Frame = -1
Query: 538 TNVAYERLPLEEDESLQMQQGQSSAGGGSGSGGGGGVNNSFPDPSS----GLPFFNLPLN 371
TNVAYERLPLEEDE Q GGGS GG N FP+ ++ GLPFFNLP+N
Sbjct: 222 TNVAYERLPLEEDEQ------QQQLGGGSNGGG-----NLFPEVAAGGGGGLPFFNLPMN 270
Query: 370 MP---QLPVDGWAGNSGGR 323
M QLPV+GW GNSGGR
Sbjct: 271 MQPNVQLPVEGWPGNSGGR 289
>ref|NP_181070.1| putative AT-hook DNA-binding protein; protein id: At2g35270.1
[Arabidopsis thaliana] gi|25348244|pir||E84766 probable
AT-hook DNA-binding protein [imported] - Arabidopsis
thaliana gi|3668079|gb|AAC61811.1| putative AT-hook
DNA-binding protein [Arabidopsis thaliana]
Length = 285
Score = 68.9 bits (167), Expect = 4e-11
Identities = 35/71 (49%), Positives = 43/71 (60%)
Frame = -1
Query: 538 TNVAYERLPLEEDESLQMQQGQSSAGGGSGSGGGGGVNNSFPDPSSGLPFFNLPLNMPQL 359
TNVAYERLPL+E E +Q G G GGGG + + GLPFFNLP++MPQ+
Sbjct: 218 TNVAYERLPLDEHEE-HLQSG--------GGGGGGNMYSEATGGGGGLPFFNLPMSMPQI 268
Query: 358 PVDGWAGNSGG 326
V+ W GN G
Sbjct: 269 GVESWQGNHAG 279
>ref|NP_566232.1| expressed protein; protein id: At3g04570.1, supported by cDNA:
15781. [Arabidopsis thaliana]
gi|6175162|gb|AAF04888.1|AC011437_3 hypothetical protein
[Arabidopsis thaliana] gi|21553701|gb|AAM62794.1|
putative DNA-binding protein [Arabidopsis thaliana]
gi|29028876|gb|AAO64817.1| At3g04570 [Arabidopsis
thaliana]
Length = 315
Score = 52.0 bits (123), Expect = 5e-06
Identities = 30/79 (37%), Positives = 41/79 (50%), Gaps = 4/79 (5%)
Frame = -1
Query: 538 TNVAYERLPLEEDESLQMQQGQSSAGGGSGSGGGGGVNNSF----PDPSSGLPFFNLPLN 371
+N YERLPLEE+E+ + G S G G GGGG S D + GLP +N+P N
Sbjct: 228 SNATYERLPLEEEEAAERGGGGGSGGVVPGQLGGGGSPLSSGAGGGDGNQGLPVYNMPGN 287
Query: 370 MPQLPVDGWAGNSGGRQSY 314
+ G G G+++Y
Sbjct: 288 LVSNGGSGGGGQMSGQEAY 306
>emb|CAC42230.1| Krueppel-like protein [Plasmodium falciparum 3D7]
Length = 1266
Score = 47.8 bits (112), Expect = 9e-05
Identities = 23/43 (53%), Positives = 29/43 (66%)
Frame = -3
Query: 269 IYIYIYIYIYIYIYIYINFLVNFLLSFELFVKKY*LLLKFVFF 141
+YIYIYIYIYIYIYIYI F+ ++ S ++ Y L F FF
Sbjct: 1210 VYIYIYIYIYIYIYIYI-FIYLYIYSLYFYIGTYSLYFIFFFF 1251
Score = 45.8 bits (107), Expect = 3e-04
Identities = 20/44 (45%), Positives = 30/44 (67%)
Frame = -3
Query: 269 IYIYIYIYIYIYIYIYINFLVNFLLSFELFVKKY*LLLKFVFFC 138
IYIYIYIYIYI+IY+YI L ++ ++ L+ + + F+F C
Sbjct: 1216 IYIYIYIYIYIFIYLYIYSLYFYIGTYSLYFIFFFFFVLFIFSC 1259
Score = 38.9 bits (89), Expect = 0.040
Identities = 16/19 (84%), Positives = 18/19 (94%)
Frame = -2
Query: 270 HLYIYIYIYIYIYIYIY*F 214
++YIYIYIYIYIYIYIY F
Sbjct: 1209 YVYIYIYIYIYIYIYIYIF 1227
Score = 38.5 bits (88), Expect = 0.052
Identities = 16/27 (59%), Positives = 22/27 (81%)
Frame = +3
Query: 204 IHQKINIYIYIYIYIYIYIYIDVKIQL 284
I++ + IYIYIYIYIYIYIYI + + +
Sbjct: 1206 IYKYVYIYIYIYIYIYIYIYIFIYLYI 1232
Score = 38.1 bits (87), Expect = 0.068
Identities = 19/40 (47%), Positives = 25/40 (62%), Gaps = 9/40 (22%)
Frame = +3
Query: 201 EIHQKIN---------IYIYIYIYIYIYIYIDVKIQLLTF 293
++ QK+N +YIYIYIYIYIYIYI + I L +
Sbjct: 1194 DVAQKVNMIYICIYKYVYIYIYIYIYIYIYIYIFIYLYIY 1233
Score = 37.7 bits (86), Expect = 0.088
Identities = 17/23 (73%), Positives = 19/23 (81%)
Frame = +1
Query: 220 IYIYIYIYIYIYIYI*MLRFNYS 288
IYIYIYIYIYIYIYI + + YS
Sbjct: 1212 IYIYIYIYIYIYIYIFIYLYIYS 1234
Score = 37.7 bits (86), Expect = 0.088
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Frame = +1
Query: 145 KTNFSNNQYFFTNNSKERRKF---TRKLIYIYIYIYIYIYIYI*MLRFNY 285
K +N FF + +++ K +YIYIYIYIYIYIYI + + Y
Sbjct: 1182 KKRITNENMFFLDVAQKVNMIYICIYKYVYIYIYIYIYIYIYIYIFIYLY 1231
Score = 37.0 bits (84), Expect = 0.15
Identities = 20/48 (41%), Positives = 25/48 (51%)
Frame = +1
Query: 142 KKTNFSNNQYFFTNNSKERRKFTRKLIYIYIYIYIYIYIYI*MLRFNY 285
KK + N +F K + Y+YIYIYIYIYIYI + F Y
Sbjct: 1182 KKRITNENMFFLDVAQKVNMIYICIYKYVYIYIYIYIYIYIYIYIFIY 1229
Score = 37.0 bits (84), Expect = 0.15
Identities = 18/26 (69%), Positives = 20/26 (76%)
Frame = +3
Query: 216 INIYIYIYIYIYIYIYIDVKIQLLTF 293
I IYIYIYIYIYIYI+I + I L F
Sbjct: 1212 IYIYIYIYIYIYIYIFIYLYIYSLYF 1237
Score = 35.4 bits (80), Expect = 0.44
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = -1
Query: 277 ILTSIYIYIYIYIYIYIYILIF 212
I IY Y+YIYIYIYIYI I+
Sbjct: 1202 IYICIYKYVYIYIYIYIYIYIY 1223
>gb|EAA13112.1| ebiP1409 [Anopheles gambiae str. PEST]
Length = 104
Score = 47.8 bits (112), Expect = 9e-05
Identities = 21/45 (46%), Positives = 31/45 (68%)
Frame = +3
Query: 162 QSVLLHKQFKGKKEIHQKINIYIYIYIYIYIYIYIDVKIQLLTFV 296
+++ +H KK+I+ I IYIYIYIYIYIYIYI + I + ++
Sbjct: 25 RNINIHTNIYNKKDIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYI 69
Score = 45.4 bits (106), Expect = 4e-04
Identities = 20/41 (48%), Positives = 27/41 (65%)
Frame = -3
Query: 269 IYIYIYIYIYIYIYIYINFLVNFLLSFELFVKKY*LLLKFV 147
IYIYIYIYIYIYIYIYI + + +++ Y L+ F+
Sbjct: 49 IYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYTYSTLMFFI 89
Score = 43.1 bits (100), Expect = 0.002
Identities = 18/43 (41%), Positives = 28/43 (64%)
Frame = -3
Query: 272 NIYIYIYIYIYIYIYIYINFLVNFLLSFELFVKKY*LLLKFVF 144
+IYIYIYIYIYIYIYIYI + + +++ Y + +++
Sbjct: 38 DIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIY 80
Score = 42.4 bits (98), Expect = 0.004
Identities = 18/37 (48%), Positives = 25/37 (66%)
Frame = -3
Query: 278 NLNIYIYIYIYIYIYIYIYINFLVNFLLSFELFVKKY 168
++ IYIYIYIYIYIYIYIYI + + +++ Y
Sbjct: 38 DIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIY 74
Score = 42.0 bits (97), Expect = 0.005
Identities = 22/43 (51%), Positives = 26/43 (60%)
Frame = -3
Query: 269 IYIYIYIYIYIYIYIYINFLVNFLLSFELFVKKY*LLLKFVFF 141
IYIYIYIYIYIYIYIYI L F + + K + F+ F
Sbjct: 61 IYIYIYIYIYIYIYIYIYIYTYSTLMFFIQLFKIKSISIFILF 103
Score = 41.2 bits (95), Expect = 0.008
Identities = 22/34 (64%), Positives = 24/34 (69%)
Frame = +1
Query: 220 IYIYIYIYIYIYIYI*MLRFNYSPLCPEDQLNKI 321
IYIYIYIYIYIYIYI + + YS L QL KI
Sbjct: 61 IYIYIYIYIYIYIYIYIYIYTYSTLMFFIQLFKI 94
Score = 41.2 bits (95), Expect = 0.008
Identities = 18/34 (52%), Positives = 23/34 (66%)
Frame = -3
Query: 269 IYIYIYIYIYIYIYIYINFLVNFLLSFELFVKKY 168
IYIYIYIYIYIYIYIYI + + F++ +
Sbjct: 59 IYIYIYIYIYIYIYIYIYIYIYTYSTLMFFIQLF 92
Score = 40.8 bits (94), Expect = 0.010
Identities = 20/36 (55%), Positives = 25/36 (68%)
Frame = +3
Query: 204 IHQKINIYIYIYIYIYIYIYIDVKIQLLTFVPRRST 311
I+ I IYIYIYIYIYIYIYI + I + ++ ST
Sbjct: 49 IYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYTYST 84
Score = 38.9 bits (89), Expect = 0.040
Identities = 19/31 (61%), Positives = 22/31 (70%)
Frame = +3
Query: 204 IHQKINIYIYIYIYIYIYIYIDVKIQLLTFV 296
I+ I IYIYIYIYIYIYIYI L+ F+
Sbjct: 59 IYIYIYIYIYIYIYIYIYIYIYTYSTLMFFI 89
Score = 37.4 bits (85), Expect = 0.12
Identities = 20/41 (48%), Positives = 23/41 (55%)
Frame = +1
Query: 142 KKTNFSNNQYFFTNNSKERRKFTRKLIYIYIYIYIYIYIYI 264
KK N TN ++ + IYIYIYIYIYIYIYI
Sbjct: 19 KKHKTYRNINIHTNIYNKKDIYIYIYIYIYIYIYIYIYIYI 59
Score = 35.0 bits (79), Expect = 0.57
Identities = 17/49 (34%), Positives = 28/49 (56%), Gaps = 4/49 (8%)
Frame = -3
Query: 278 NLNIYIYIY----IYIYIYIYIYINFLVNFLLSFELFVKKY*LLLKFVF 144
N+NI+ IY IYIYIYIYIYI + + +++ Y + +++
Sbjct: 26 NINIHTNIYNKKDIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIYIY 74
Score = 32.0 bits (71), Expect = 4.8
Identities = 16/38 (42%), Positives = 24/38 (63%), Gaps = 4/38 (10%)
Frame = +3
Query: 195 KKEIHQKINIYIYIY----IYIYIYIYIDVKIQLLTFV 296
K + ++ INI+ IY IYIYIYIYI + I + ++
Sbjct: 20 KHKTYRNINIHTNIYNKKDIYIYIYIYIYIYIYIYIYI 57
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 466,621,749
Number of Sequences: 1393205
Number of extensions: 12145434
Number of successful extensions: 196145
Number of sequences better than 10.0: 646
Number of HSP's better than 10.0 without gapping: 65745
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 144622
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18173652336
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)