Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002307A_C01 KMC002307A_c01
(578 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
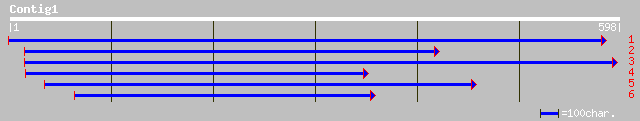
dbj|BAB64937.1| TdcA1-ORF1-ORF2 [Daucus carota] 43 0.003
gb|AAL68772.1|AF445084_1 putative transposase A [Lactobacillus c... 37 0.23
emb|CAB63122.1| transposase [Lactobacillus sanfranciscensis] 36 0.30
ref|NP_200770.1| putative protein; protein id: At5g59620.1 [Arab... 36 0.40
gb|EAA21227.1| Ubiquitin family, putative [Plasmodium yoelii yoe... 34 1.2
>dbj|BAB64937.1| TdcA1-ORF1-ORF2 [Daucus carota]
Length = 988
Score = 42.7 bits (99), Expect = 0.003
Identities = 30/79 (37%), Positives = 44/79 (54%), Gaps = 7/79 (8%)
Frame = -3
Query: 519 ENEDGAHKKSNDEIYLEVVGGANKKGRIYGLGA-EAVKF----KRSKATSSLGVS--PSE 361
E D DEI+LE VGG +K+ RIYGLG+ ++V + K S +TS S E
Sbjct: 879 EGSDEPQIVDEDEIFLEAVGGLDKRNRIYGLGSLQSVIYGPESKSSTSTSRYSGSNFNKE 938
Query: 360 YETMRNLVSTLTEENKKLK 304
YE M+ + + E+ K+L+
Sbjct: 939 YELMQVELQEMKEQVKELQ 957
>gb|AAL68772.1|AF445084_1 putative transposase A [Lactobacillus casei]
Length = 83
Score = 36.6 bits (83), Expect = 0.23
Identities = 21/61 (34%), Positives = 35/61 (56%), Gaps = 1/61 (1%)
Frame = -3
Query: 456 ANKKGRIYGLGAEAV-KFKRSKATSSLGVSPSEYETMRNLVSTLTEENKKLKGKMGTYDE 280
A + R YG+G V K+ + +A + G SP E + M +++L+EEN+ LK +G +
Sbjct: 23 AAQMAREYGIGYSTVHKWIQGQAKTQSGKSPDEIKAMEKRLASLSEENEILKKALGFLAQ 82
Query: 279 K 277
K
Sbjct: 83 K 83
>emb|CAB63122.1| transposase [Lactobacillus sanfranciscensis]
Length = 83
Score = 36.2 bits (82), Expect = 0.30
Identities = 21/61 (34%), Positives = 35/61 (56%), Gaps = 1/61 (1%)
Frame = -3
Query: 456 ANKKGRIYGLGAEAV-KFKRSKATSSLGVSPSEYETMRNLVSTLTEENKKLKGKMGTYDE 280
A + R YG+G V K+ + +A + G SP E + M +++L+EEN+ LK +G +
Sbjct: 23 AAQLAREYGIGYSTVHKWIQGQAKTQSGKSPDEIKAMEKRLASLSEENEILKKALGFLAQ 82
Query: 279 K 277
K
Sbjct: 83 K 83
>ref|NP_200770.1| putative protein; protein id: At5g59620.1 [Arabidopsis thaliana]
gi|9758829|dbj|BAB09501.1| contains similarity to
En/Spm-like transposon protein~gene_id:MTH12.16
[Arabidopsis thaliana]
Length = 312
Score = 35.8 bits (81), Expect = 0.40
Identities = 24/86 (27%), Positives = 45/86 (51%)
Frame = -3
Query: 501 HKKSNDEIYLEVVGGANKKGRIYGLGAEAVKFKRSKATSSLGVSPSEYETMRNLVSTLTE 322
H+K+ EIY++ G ++K G I+GLGA A+SS + E ET+ + + +
Sbjct: 225 HEKN--EIYIKAAG-SSKHGHIFGLGALMETLPSVGASSSAPQASEEVETITHRLQEMET 281
Query: 321 ENKKLKGKMGTYDEKFANLDRFLETF 244
+ KK + ++ +++ +ETF
Sbjct: 282 DLKKSLEENLQIQKRLEAMEKLVETF 307
>gb|EAA21227.1| Ubiquitin family, putative [Plasmodium yoelii yoelii]
Length = 1326
Score = 34.3 bits (77), Expect = 1.2
Identities = 36/129 (27%), Positives = 55/129 (41%), Gaps = 8/129 (6%)
Frame = -3
Query: 537 AARDSQENEDGAHKKSNDEIYLEVVGGANKKGRIYGLGAEAVKFKRSKATSSLGVSP-SE 361
A+ D+ +N D +N++ G N K G K KR+ ++SS S S
Sbjct: 300 ASNDNNDNNDDNGSGNNNDANGNGNGNGNNKHSREGRN----KGKRNNSSSSSSSSENSS 355
Query: 360 YETMRNLVSTLTEENKKLKGKM-GTYDEKFANL------DRFLETFP*SMASRSVEQSPT 202
YE +N + E+ KKLK K+ Y EK N D + ++ S E S
Sbjct: 356 YEYNKNKKESDEEKKKKLKKKLKKKYMEKIRNKYSENINDIYSSSYSSDYESSYSEISDR 415
Query: 201 NQHNIDDEE 175
+ +DDE+
Sbjct: 416 KERELDDEK 424
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 470,562,584
Number of Sequences: 1393205
Number of extensions: 9755107
Number of successful extensions: 34890
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 32806
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34767
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21426319650
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)