Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002299A_C01 KMC002299A_c01
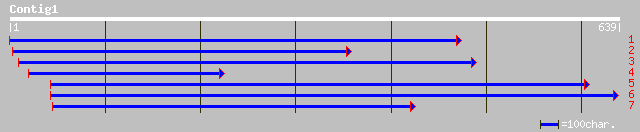
(636 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB90157.1| P0408G07.3 [Oryza sativa (japonica cultivar-group)] 186 2e-46
ref|NP_564182.1| expressed protein; protein id: At1g22850.1, sup... 184 1e-45
ref|ZP_00073678.1| hypothetical protein [Trichodesmium erythraeu... 77 2e-13
gb|ZP_00110031.1| hypothetical protein [Nostoc punctiforme] 75 6e-13
ref|NP_486247.1| hypothetical protein [Nostoc sp. PCC 7120] gi|2... 72 6e-12
>dbj|BAB90157.1| P0408G07.3 [Oryza sativa (japonica cultivar-group)]
Length = 340
Score = 186 bits (472), Expect = 2e-46
Identities = 93/104 (89%), Positives = 101/104 (96%)
Frame = -2
Query: 635 NGFKVVTLLRLSPLLPFSLGNYLYGLTSVKFLPYVLGSWLGMLPGTWAYVSAGAFGRAII 456
NGFKVVTLLRLSPLLPFSLGNYLYGLTSVKFLPYVLGSWLGMLPG+WAYVSAGAFGRAII
Sbjct: 237 NGFKVVTLLRLSPLLPFSLGNYLYGLTSVKFLPYVLGSWLGMLPGSWAYVSAGAFGRAII 296
Query: 455 QEESELKVLGGNSQLLTLGLGLLVTALAATYVTRLAKDAIKDIE 324
Q+ESE+ LGGNSQLLTLG+GLL TA+AATYVTRLAKDA+K+I+
Sbjct: 297 QDESEIG-LGGNSQLLTLGIGLLFTAIAATYVTRLAKDAVKEID 339
>ref|NP_564182.1| expressed protein; protein id: At1g22850.1, supported by cDNA:
gi_15215601, supported by cDNA: gi_20856178 [Arabidopsis
thaliana] gi|25518503|pir||D86362 hypothetical protein
F29G20.19 - Arabidopsis thaliana
gi|2462839|gb|AAB72174.1| unknown protein [Arabidopsis
thaliana] gi|15215602|gb|AAK91346.1| At1g22850/F29G20_19
[Arabidopsis thaliana] gi|20856179|gb|AAM26652.1|
At1g22850/F29G20_19 [Arabidopsis thaliana]
Length = 344
Score = 184 bits (466), Expect = 1e-45
Identities = 91/104 (87%), Positives = 98/104 (93%)
Frame = -2
Query: 635 NGFKVVTLLRLSPLLPFSLGNYLYGLTSVKFLPYVLGSWLGMLPGTWAYVSAGAFGRAII 456
NGF+VVTLLRLSPLLPFSLGNYLYGLTSVKF+PYVLGSWLGMLPG+WAYVSAGAFGRAII
Sbjct: 233 NGFRVVTLLRLSPLLPFSLGNYLYGLTSVKFVPYVLGSWLGMLPGSWAYVSAGAFGRAII 292
Query: 455 QEESELKVLGGNSQLLTLGLGLLVTALAATYVTRLAKDAIKDIE 324
QEES + + GGN QLLTLG+GLLVTALA TYVT LAKDAIKDI+
Sbjct: 293 QEESNVGLPGGNGQLLTLGVGLLVTALAGTYVTSLAKDAIKDID 336
>ref|ZP_00073678.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 242
Score = 77.0 bits (188), Expect = 2e-13
Identities = 39/100 (39%), Positives = 60/100 (60%)
Frame = -2
Query: 632 GFKVVTLLRLSPLLPFSLGNYLYGLTSVKFLPYVLGSWLGMLPGTWAYVSAGAFGRAIIQ 453
G+K+V L RLSP+ PF+L NY +GLT V Y SW+GM+PGT YV G+ ++
Sbjct: 140 GWKIVGLTRLSPIFPFNLLNYAFGLTQVSLQHYFFASWIGMMPGTVMYVYLGSLAGSLAT 199
Query: 452 EESELKVLGGNSQLLTLGLGLLVTALAATYVTRLAKDAIK 333
+E + ++ + G+GL+ T YVT++AK A++
Sbjct: 200 LGTEER-SRTTTEWVLYGVGLIATVAVTFYVTKIAKKALQ 238
>gb|ZP_00110031.1| hypothetical protein [Nostoc punctiforme]
Length = 256
Score = 75.5 bits (184), Expect = 6e-13
Identities = 40/101 (39%), Positives = 58/101 (56%)
Frame = -2
Query: 632 GFKVVTLLRLSPLLPFSLGNYLYGLTSVKFLPYVLGSWLGMLPGTWAYVSAGAFGRAIIQ 453
G K+V L RLSP+ PF+L NY +G+T V Y +GS LGM+PGT YV G+ +
Sbjct: 155 GLKIVLLTRLSPIFPFNLLNYAFGITGVSLKDYFIGS-LGMIPGTIMYVYIGSLASNLAM 213
Query: 452 EESELKVLGGNSQLLTLGLGLLVTALAATYVTRLAKDAIKD 330
+E ++ Q LGL+ T YVTR+A+ A+++
Sbjct: 214 IGTEAQLTNPTLQWAIRILGLIATVAVTVYVTRIARKALEE 254
>ref|NP_486247.1| hypothetical protein [Nostoc sp. PCC 7120] gi|25326049|pir||AI2081
hypothetical protein alr2207 [imported] - Nostoc sp.
(strain PCC 7120) gi|17131298|dbj|BAB73906.1|
ORF_ID:alr2207~hypothetical protein [Nostoc sp. PCC
7120]
Length = 282
Score = 72.0 bits (175), Expect = 6e-12
Identities = 40/101 (39%), Positives = 57/101 (55%)
Frame = -2
Query: 632 GFKVVTLLRLSPLLPFSLGNYLYGLTSVKFLPYVLGSWLGMLPGTWAYVSAGAFGRAIIQ 453
G K+V L RLSP+ PF+L NY YG+T V YVL S +GM+PGT YV G+ +I
Sbjct: 166 GLKIVLLTRLSPIFPFNLLNYAYGVTGVSLKDYVLAS-IGMIPGTIMYVYIGSLAGSIAT 224
Query: 452 EESELKVLGGNSQLLTLGLGLLVTALAATYVTRLAKDAIKD 330
+E + Q +G + T YVT++A+ A++D
Sbjct: 225 IGTESQPGNPGVQWAIRIIGFIATVAVTIYVTKVARKALED 265
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 527,476,244
Number of Sequences: 1393205
Number of extensions: 11251510
Number of successful extensions: 30714
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 29358
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30678
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)