Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002292A_C01 KMC002292A_c01
(572 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
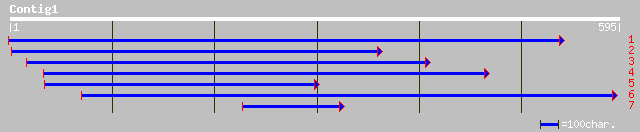
ref|NP_197266.1| putative protein; protein id: At5g17640.1 [Arab... 132 4e-30
dbj|BAC65069.1| OJ9990_A01.12 [Oryza sativa (japonica cultivar-g... 96 2e-19
ref|NP_172473.1| unknown protein; protein id: At1g10020.1 [Arabi... 82 4e-15
ref|NP_194660.1| putative protein; protein id: At4g29310.1, supp... 80 1e-14
gb|AAL60032.1| unknown protein [Arabidopsis thaliana] 77 1e-13
>ref|NP_197266.1| putative protein; protein id: At5g17640.1 [Arabidopsis thaliana]
gi|11282612|pir||T51468 hypothetical protein K10A8_120 -
Arabidopsis thaliana gi|9755789|emb|CAC01908.1| putative
protein [Arabidopsis thaliana]
Length = 432
Score = 132 bits (331), Expect = 4e-30
Identities = 68/95 (71%), Positives = 75/95 (78%), Gaps = 2/95 (2%)
Frame = -3
Query: 570 DTEKHMRNAAASPVPSPQSSGDFGALSPVV--GGFVMCCRVQGEGKRGKPLVQLALRHVT 397
DT+K M AA+P+PSPQSSGDF L V GGFVM RVQGEGK KP+VQLA+RHVT
Sbjct: 335 DTDKQMLTVAATPIPSPQSSGDFSGLGQCVSGGGFVMSSRVQGEGKSSKPVVQLAMRHVT 394
Query: 396 CVEDAAIFMALAAAVDLSIEACRPFRRKIRRGKQH 292
CVEDAAIFMALAAAVDLSI AC+PFRR RR +H
Sbjct: 395 CVEDAAIFMALAAAVDLSILACKPFRRTSRRRFRH 429
>dbj|BAC65069.1| OJ9990_A01.12 [Oryza sativa (japonica cultivar-group)]
Length = 439
Score = 96.3 bits (238), Expect = 2e-19
Identities = 44/62 (70%), Positives = 54/62 (86%)
Frame = -3
Query: 480 GGFVMCCRVQGEGKRGKPLVQLALRHVTCVEDAAIFMALAAAVDLSIEACRPFRRKIRRG 301
GGFVM CRV+GE + +PLVQLA+RHVTC+EDAA+F+ALAAAVDLS++ACRPFRRK
Sbjct: 363 GGFVMSCRVEGESRSSRPLVQLAMRHVTCMEDAAMFVALAAAVDLSVKACRPFRRKTTAT 422
Query: 300 KQ 295
K+
Sbjct: 423 KK 424
>ref|NP_172473.1| unknown protein; protein id: At1g10020.1 [Arabidopsis thaliana]
gi|7487490|pir||T00621 hypothetical protein T27I1.4 -
Arabidopsis thaliana gi|3540181|gb|AAC34331.1| Unknown
protein [Arabidopsis thaliana]
Length = 461
Score = 82.4 bits (202), Expect = 4e-15
Identities = 44/87 (50%), Positives = 64/87 (72%), Gaps = 4/87 (4%)
Frame = -3
Query: 540 ASPVPSPQS-SGDFG-ALSP--VVGGFVMCCRVQGEGKRGKPLVQLALRHVTCVEDAAIF 373
ASP SP+ SGD+G L P V GFVM V+GEGK KP V+++++HV+C+EDAA +
Sbjct: 371 ASPANSPRGGSGDYGYGLWPWNVYKGFVMSASVEGEGKCSKPCVEVSVQHVSCMEDAAAY 430
Query: 372 MALAAAVDLSIEACRPFRRKIRRGKQH 292
+AL+AA+DLS++ACR F +++R+ H
Sbjct: 431 VALSAAIDLSMDACRLFNQRMRKELCH 457
>ref|NP_194660.1| putative protein; protein id: At4g29310.1, supported by cDNA:
gi_20260629 [Arabidopsis thaliana]
gi|7487111|pir||T13451 hypothetical protein T17A13.130 -
Arabidopsis thaliana gi|7269829|emb|CAB79689.1| putative
protein [Arabidopsis thaliana]
gi|20260630|gb|AAM13213.1| unknown protein [Arabidopsis
thaliana] gi|28059515|gb|AAO30065.1| unknown protein
[Arabidopsis thaliana]
Length = 424
Score = 80.5 bits (197), Expect = 1e-14
Identities = 39/73 (53%), Positives = 54/73 (73%)
Frame = -3
Query: 510 GDFGALSPVVGGFVMCCRVQGEGKRGKPLVQLALRHVTCVEDAAIFMALAAAVDLSIEAC 331
G+ A+S V GFVM V+GEGK KP+V + +HVTC+ DAA+F+AL+AAVDLS++AC
Sbjct: 345 GESPAISSPVKGFVMGSSVEGEGKVSKPVVHVGAQHVTCMADAALFVALSAAVDLSVDAC 404
Query: 330 RPFRRKIRRGKQH 292
+ F RK+R+ H
Sbjct: 405 QLFSRKLRKELCH 417
>gb|AAL60032.1| unknown protein [Arabidopsis thaliana]
Length = 491
Score = 77.4 bits (189), Expect = 1e-13
Identities = 42/82 (51%), Positives = 51/82 (61%), Gaps = 7/82 (8%)
Frame = -3
Query: 528 PSPQSSGDFGALSPVVG-------GFVMCCRVQGEGKRGKPLVQLALRHVTCVEDAAIFM 370
P S DFG L P GFVM V+G GKR KP V++ + HVTC EDAA +
Sbjct: 399 PGSGSGSDFGYLLPQHPSAAAQNRGFVMSATVEGVGKRSKPEVEVGVTHVTCTEDAAAHV 458
Query: 369 ALAAAVDLSIEACRPFRRKIRR 304
ALAAAVDLS++ACR F K+R+
Sbjct: 459 ALAAAVDLSLDACRLFSHKLRK 480
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 472,925,661
Number of Sequences: 1393205
Number of extensions: 9772743
Number of successful extensions: 29381
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 27877
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29265
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)