Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
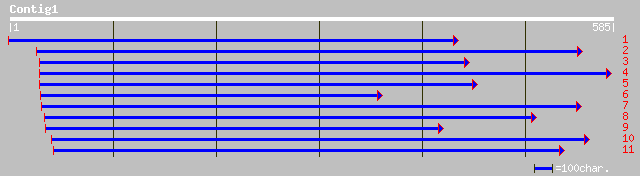
Query= KMC002289A_C01 KMC002289A_c01
(580 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_568928.1| putative protein; protein id: At5g60750.1, supp... 176 2e-43
dbj|BAB09848.1| gene_id:MUP24.17~pir||S76167~similar to unknown ... 176 2e-43
ref|NP_487825.1| hypothetical protein [Nostoc sp. PCC 7120] gi|2... 117 1e-30
gb|ZP_00111909.1| hypothetical protein [Nostoc punctiforme] 117 3e-30
ref|ZP_00074521.1| hypothetical protein [Trichodesmium erythraeu... 125 4e-28
>ref|NP_568928.1| putative protein; protein id: At5g60750.1, supported by cDNA:
25855., supported by cDNA: gi_17528993, supported by
cDNA: gi_20465442 [Arabidopsis thaliana]
gi|17528994|gb|AAL38707.1| unknown protein [Arabidopsis
thaliana] gi|20465443|gb|AAM20181.1| unknown protein
[Arabidopsis thaliana] gi|21554615|gb|AAM63634.1|
unknown [Arabidopsis thaliana]
Length = 347
Score = 176 bits (446), Expect = 2e-43
Identities = 80/96 (83%), Positives = 92/96 (95%)
Frame = -3
Query: 341 AMALYAVVVSVCAPIWEEIVFRGFLLPSLTRYMPVWSAILVSSVAFALAHFNIQRMIPLI 162
AMALYAVVVS+CAP+WEEIVFRGFLLPSLTRYMPVW AILVSS+AFALAHFN+QRM+PL+
Sbjct: 252 AMALYAVVVSICAPVWEEIVFRGFLLPSLTRYMPVWCAILVSSIAFALAHFNVQRMLPLV 311
Query: 161 FLGIVMGAVFVRSRNLLPSMLLHSLWNAFVFLDLMK 54
FLG+V+G +F RSRNLLPSMLLHSLWN FVF++LM+
Sbjct: 312 FLGVVLGLIFARSRNLLPSMLLHSLWNGFVFMELMR 347
Score = 112 bits (279), Expect = 4e-24
Identities = 57/101 (56%), Positives = 75/101 (73%), Gaps = 1/101 (0%)
Frame = -2
Query: 579 GVVGIAILRRCLASFQPLPPNWFKFELKGNWQFDVGLGCLMFPLINQLSQMNLNLLPV-L 403
G+ GIAIL RCL+ F+PL +WF+F LKGNWQ DV +GC MFP +N+LSQ+NLNLLP+
Sbjct: 172 GLAGIAILHRCLSMFRPLASDWFRFTLKGNWQLDVIIGCFMFPFVNRLSQLNLNLLPLPP 231
Query: 402 QYTPVTVSSVEQSIVARDPVRYGPLCGGGLSLCADMGGDCV 280
+PV++SSVEQSI+ARDPV L +S+CA + + V
Sbjct: 232 TSSPVSLSSVEQSIMARDPVAMA-LYAVVVSICAPVWEEIV 271
>dbj|BAB09848.1| gene_id:MUP24.17~pir||S76167~similar to unknown protein
[Arabidopsis thaliana]
Length = 312
Score = 176 bits (446), Expect = 2e-43
Identities = 80/96 (83%), Positives = 92/96 (95%)
Frame = -3
Query: 341 AMALYAVVVSVCAPIWEEIVFRGFLLPSLTRYMPVWSAILVSSVAFALAHFNIQRMIPLI 162
AMALYAVVVS+CAP+WEEIVFRGFLLPSLTRYMPVW AILVSS+AFALAHFN+QRM+PL+
Sbjct: 217 AMALYAVVVSICAPVWEEIVFRGFLLPSLTRYMPVWCAILVSSIAFALAHFNVQRMLPLV 276
Query: 161 FLGIVMGAVFVRSRNLLPSMLLHSLWNAFVFLDLMK 54
FLG+V+G +F RSRNLLPSMLLHSLWN FVF++LM+
Sbjct: 277 FLGVVLGLIFARSRNLLPSMLLHSLWNGFVFMELMR 312
Score = 112 bits (279), Expect = 4e-24
Identities = 57/101 (56%), Positives = 75/101 (73%), Gaps = 1/101 (0%)
Frame = -2
Query: 579 GVVGIAILRRCLASFQPLPPNWFKFELKGNWQFDVGLGCLMFPLINQLSQMNLNLLPV-L 403
G+ GIAIL RCL+ F+PL +WF+F LKGNWQ DV +GC MFP +N+LSQ+NLNLLP+
Sbjct: 137 GLAGIAILHRCLSMFRPLASDWFRFTLKGNWQLDVIIGCFMFPFVNRLSQLNLNLLPLPP 196
Query: 402 QYTPVTVSSVEQSIVARDPVRYGPLCGGGLSLCADMGGDCV 280
+PV++SSVEQSI+ARDPV L +S+CA + + V
Sbjct: 197 TSSPVSLSSVEQSIMARDPVAMA-LYAVVVSICAPVWEEIV 236
>ref|NP_487825.1| hypothetical protein [Nostoc sp. PCC 7120] gi|25365094|pir||AB2279
hypothetical protein all3785 [imported] - Nostoc sp.
(strain PCC 7120) gi|17132919|dbj|BAB75484.1|
ORF_ID:all3785~hypothetical protein [Nostoc sp. PCC
7120]
Length = 528
Score = 117 bits (294), Expect(2) = 1e-30
Identities = 52/94 (55%), Positives = 74/94 (78%)
Frame = -3
Query: 341 AMALYAVVVSVCAPIWEEIVFRGFLLPSLTRYMPVWSAILVSSVAFALAHFNIQRMIPLI 162
A++++ V ++ AP++EE++FRGFLLPSLTRY+PVW AI+ S+V FA+AH ++ ++PL
Sbjct: 428 ALSIFYVTAAIAAPLFEEVLFRGFLLPSLTRYIPVWGAIIASAVLFAVAHLSLSEILPLT 487
Query: 161 FLGIVMGAVFVRSRNLLPSMLLHSLWNAFVFLDL 60
LGIV+G V+ RSRNLL +LLHSLWN+ L L
Sbjct: 488 ALGIVLGVVYTRSRNLLAPILLHSLWNSGTLLSL 521
Score = 37.0 bits (84), Expect(2) = 1e-30
Identities = 20/53 (37%), Positives = 30/53 (55%), Gaps = 2/53 (3%)
Frame = -2
Query: 567 IAILRRCLASFQPLPPNWFKFELKGNWQFDVGLG--CLMFPLINQLSQMNLNL 415
+++L + F PLP WF+F L+ NW F GLG C P++ +S +N L
Sbjct: 355 LSVLYFSIKRFFPLPQYWFRFRLRDNW-FLWGLGGYCAALPIVVIVSLINQQL 406
>gb|ZP_00111909.1| hypothetical protein [Nostoc punctiforme]
Length = 524
Score = 117 bits (293), Expect(2) = 3e-30
Identities = 52/94 (55%), Positives = 72/94 (76%)
Frame = -3
Query: 341 AMALYAVVVSVCAPIWEEIVFRGFLLPSLTRYMPVWSAILVSSVAFALAHFNIQRMIPLI 162
A+ ++ ++ AP +EEI+FRGFLLPSLTRY+PVW +I++SS+ FA+AH ++ ++PL
Sbjct: 425 ALGIFFFTAAIAAPFFEEILFRGFLLPSLTRYLPVWGSIIISSLLFAIAHLSLSEILPLT 484
Query: 161 FLGIVMGAVFVRSRNLLPSMLLHSLWNAFVFLDL 60
LGIV+G V+ RSRNLL MLLHSLWN+ L L
Sbjct: 485 ALGIVLGVVYTRSRNLLAPMLLHSLWNSGTLLSL 518
Score = 36.2 bits (82), Expect(2) = 3e-30
Identities = 20/53 (37%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Frame = -2
Query: 567 IAILRRCLASFQPLPPNWFKFELKGNWQFDVGLG--CLMFPLINQLSQMNLNL 415
+ +L L F PLP WF+F + NW F GLG C P++ +S +N L
Sbjct: 352 LLVLYFSLKPFFPLPEFWFRFRFQNNW-FLWGLGGYCTALPIVVMVSLINQQL 403
>ref|ZP_00074521.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 119
Score = 125 bits (314), Expect = 4e-28
Identities = 56/94 (59%), Positives = 74/94 (78%)
Frame = -3
Query: 341 AMALYAVVVSVCAPIWEEIVFRGFLLPSLTRYMPVWSAILVSSVAFALAHFNIQRMIPLI 162
A+ L+ V S+ AP++EE++FRGFLLPSLT+YMP+W AI+VS FA+AH NI ++PL
Sbjct: 18 ALGLFFVTASIAAPVFEEVMFRGFLLPSLTKYMPIWGAIIVSGFLFAIAHLNISEVLPLA 77
Query: 161 FLGIVMGAVFVRSRNLLPSMLLHSLWNAFVFLDL 60
LGI++GAV+ RSRNLL S+LLHSLWN+ L L
Sbjct: 78 VLGIILGAVYTRSRNLLSSILLHSLWNSGTLLSL 111
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 508,864,639
Number of Sequences: 1393205
Number of extensions: 11195024
Number of successful extensions: 28703
Number of sequences better than 10.0: 244
Number of HSP's better than 10.0 without gapping: 27544
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28647
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21426319650
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)