Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002273A_C02 KMC002273A_c02
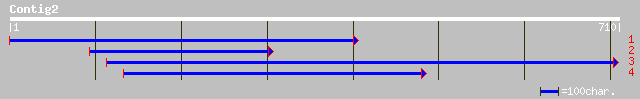
(710 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM70491.1|AF517943_1 hedgehog-like protein [Helobdella robusta] 37 0.21
dbj|BAC39198.1| unnamed protein product [Mus musculus] 34 1.8
sp|Q9RGR9|GALT_STACA Galactose-1-phosphate uridylyltransferase (... 34 2.3
ref|NP_510654.1| Predicted CDS, TWiK family of potassium channel... 33 3.0
gb|AAA31881.1| A 'c' was inserted after nt 369 (=nt 10459 in gen... 33 3.0
>gb|AAM70491.1|AF517943_1 hedgehog-like protein [Helobdella robusta]
Length = 1073
Score = 37.4 bits (85), Expect = 0.21
Identities = 26/97 (26%), Positives = 44/97 (44%), Gaps = 9/97 (9%)
Frame = +2
Query: 422 RKNLPHKLKQCAVKDLHEISSPLQCSFHNS---------ISSSSHLHLSAIIINXCQFIA 574
+K P KL+ +D+H+ Q H+ +S+S LHLS I +
Sbjct: 82 QKQQPLKLQTQKYQDIHQQQPQNQQQQHHQQQQQHLLHHLSTSLSLHLSDI---SDSYSN 138
Query: 575 EFNSTTNKTYXLNSTLQENHLNTYADTIASPHHAYHH 685
FN++ + + NH N Y D+I++ HH +H+
Sbjct: 139 AFNTSITDSSKSSPDHSTNHYNHYHDSISTHHHRHHY 175
>dbj|BAC39198.1| unnamed protein product [Mus musculus]
Length = 124
Score = 34.3 bits (77), Expect = 1.8
Identities = 19/49 (38%), Positives = 23/49 (46%)
Frame = -2
Query: 265 FLKSFLPEDGGISLVYGSCIAVFFVALILSYFLHFLTCILFSWLLSFLL 119
FL SF P S ++ +FF L LS FL F S+ LSF L
Sbjct: 58 FLPSFFPSFTFFSFIFSFLYFLFFFFLSLSLFLSFFLSFFLSFFLSFFL 106
>sp|Q9RGR9|GALT_STACA Galactose-1-phosphate uridylyltransferase (Gal-1-P
uridylyltransferase) gi|6707011|gb|AAF25550.1|AF109295_5
GalT [Staphylococcus carnosus]
Length = 498
Score = 33.9 bits (76), Expect = 2.3
Identities = 19/45 (42%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Frame = +2
Query: 569 IAEFNSTTNKTYXLNSTLQENHLNT-YADTIASPHHAYHHFVHYN 700
IA FN TT++ Y L+ L++N +T Y D I PH+ HH N
Sbjct: 360 IARFNQTTSE-YELDLVLRDNQTSTQYPDGIFHPHNDVHHIKKEN 403
>ref|NP_510654.1| Predicted CDS, TWiK family of potassium channels TWK-16 (twk-16)
[Caenorhabditis elegans] gi|7506819|pir||T24265
hypothetical protein T01B4.1 - Caenorhabditis elegans
gi|3879242|emb|CAA93875.1| C. elegans TWK-16 protein
(corresponding sequence T01B4.1) [Caenorhabditis
elegans]
Length = 522
Score = 33.5 bits (75), Expect = 3.0
Identities = 19/68 (27%), Positives = 33/68 (47%), Gaps = 2/68 (2%)
Frame = -1
Query: 371 LIILLSAFIPSWWLPLDFRNWYYFSFTGFFLVRLPFSKVFP--PRRWWDIISIWFMYCCI 198
L ++L +F+ S+W DF +YF F L + F + P PR + ++F+ +
Sbjct: 244 LFVVLCSFVVSFWENWDFLTAFYFFFVS--LSTIGFGDIVPDHPRTACALFVLYFIGLAL 301
Query: 197 FCRTYLVL 174
F Y +L
Sbjct: 302 FAMVYAIL 309
>gb|AAA31881.1| A 'c' was inserted after nt 369 (=nt 10459 in genomic sequence
(M10126)) to correct -1 frameshift probably due to gel
compression
Length = 355
Score = 33.5 bits (75), Expect = 3.0
Identities = 18/84 (21%), Positives = 38/84 (44%)
Frame = -3
Query: 363 SLICFYSFLVVTFRF*KLVLFFLHWVFPSKVAFF*SLSSQKMVGYH*YMVHVLLYFLSHL 184
+ IC++ + + + + L ++F ++F S ++ Y + + F S+L
Sbjct: 101 NFICYFFCFSIALYYIEFFTYLLCFIFIDFISF-----SNHLISYFGIINMFNVIFCSYL 155
Query: 183 SCLISFISLLVFCSLGFFRSCYYL 112
CL FI +FC + F C ++
Sbjct: 156 FCLFYFIICFIFCFIFFVIRCLFV 179
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 643,837,856
Number of Sequences: 1393205
Number of extensions: 14506655
Number of successful extensions: 42872
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 39569
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42692
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32654539052
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)