Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002271A_C01 KMC002271A_c01
(537 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
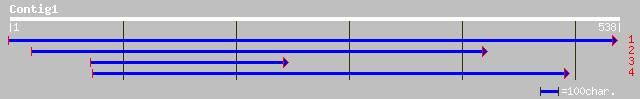
ref|NP_190382.1| putative protein; protein id: At3g47990.1 [Arab... 73 2e-12
dbj|BAC42060.1| unknown protein [Arabidopsis thaliana] 72 3e-12
gb|EAA06368.1| agCP13358 [Anopheles gambiae str. PEST] 37 0.20
ref|NP_232505.1| ribosomal large subunit pseudouridine synthase ... 34 0.98
ref|NP_353109.1| AGR_C_112p [Agrobacterium tumefaciens] gi|17933... 34 1.3
>ref|NP_190382.1| putative protein; protein id: At3g47990.1 [Arabidopsis thaliana]
gi|7487119|pir||T06684 hypothetical protein T17F15.140 -
Arabidopsis thaliana gi|4678329|emb|CAB41140.1| putative
protein [Arabidopsis thaliana]
Length = 292
Score = 72.8 bits (177), Expect = 2e-12
Identities = 41/80 (51%), Positives = 50/80 (62%), Gaps = 3/80 (3%)
Frame = -3
Query: 535 CSVFPNLDLSALSNLRADSERSPASVMTTT---RYVRGQPASQSYRLRLQGLLRPVRAEV 365
CSVFP+LDLSALSNL++ + V T T RY+R QP S+SY LR+Q L+ PV
Sbjct: 211 CSVFPDLDLSALSNLQSSGTEQHSQVNTETSEARYIRSQPQSESYFLRVQSLIHPVH--- 267
Query: 364 AGPVGDTDDALDNAENGAVP 305
TD AL+ AENG VP
Sbjct: 268 ------TDTALETAENGGVP 281
>dbj|BAC42060.1| unknown protein [Arabidopsis thaliana]
Length = 358
Score = 72.4 bits (176), Expect = 3e-12
Identities = 41/80 (51%), Positives = 50/80 (62%), Gaps = 3/80 (3%)
Frame = -3
Query: 535 CSVFPNLDLSALSNLRADSERSPASVMTTT---RYVRGQPASQSYRLRLQGLLRPVRAEV 365
CSVFP+LDLSALSNL++ + V T T RY+R QP S+SY LR+Q L+ PV A
Sbjct: 277 CSVFPDLDLSALSNLQSSGTEQHSQVNTETSEARYIRSQPQSESYFLRVQSLIHPVHA-- 334
Query: 364 AGPVGDTDDALDNAENGAVP 305
D AL+ AENG VP
Sbjct: 335 -------DTALETAENGGVP 347
>gb|EAA06368.1| agCP13358 [Anopheles gambiae str. PEST]
Length = 580
Score = 36.6 bits (83), Expect = 0.20
Identities = 19/70 (27%), Positives = 40/70 (57%)
Frame = -3
Query: 514 DLSALSNLRADSERSPASVMTTTRYVRGQPASQSYRLRLQGLLRPVRAEVAGPVGDTDDA 335
D A S+ + ++RSP+S +++ G+P ++ RL+ LR R+ V + ++ ++
Sbjct: 477 DGEATSSSSSSAKRSPSSTASSSTLPAGEPLMRTPERRLKLTLRMKRSPVIDEIIESGNS 536
Query: 334 LDNAENGAVP 305
+ +A+NG P
Sbjct: 537 MSDADNGGSP 546
>ref|NP_232505.1| ribosomal large subunit pseudouridine synthase A [Vibrio cholerae]
gi|11356080|pir||C82498 ribosomal large chain
pseudouridine synthase A VCA0104 [imported] - Vibrio
cholerae (strain N16961 serogroup O1)
gi|9657491|gb|AAF96018.1| ribosomal large subunit
pseudouridine synthase A [Vibrio cholerae]
Length = 221
Score = 34.3 bits (77), Expect = 0.98
Identities = 21/59 (35%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Frame = +1
Query: 235 RKHSQACIQQKRPAL*LMHFDSRQVQ---LRSQHYPKHHRCHQLALQLQHALGGEDPVI 402
R + C QQ +PAL H+++ QV+ R YP R HQL + H LG P++
Sbjct: 127 RPRQKVCWQQGKPAL--THWETVQVEQNRTRVYLYPHTGRTHQLRVHCAHHLGLNAPIV 183
>ref|NP_353109.1| AGR_C_112p [Agrobacterium tumefaciens] gi|17933994|ref|NP_530784.1|
conserved hypothetical protein [Agrobacterium
tumefaciens str. C58 (U. Washington)]
gi|25520214|pir||E97367 hypothetical protein AGR_C_112
[imported] - Agrobacterium tumefaciens (strain C58,
Cereon) gi|25522944|pir||AF2585 conserved hypothetical
protein Atu0075 [imported] - Agrobacterium tumefaciens
(strain C58, Dupont) gi|15154939|gb|AAK85894.1|
AGR_C_112p [Agrobacterium tumefaciens str. C58 (Cereon)]
gi|17738391|gb|AAL41100.1| conserved hypothetical
protein [Agrobacterium tumefaciens str. C58 (U.
Washington)]
Length = 158
Score = 33.9 bits (76), Expect = 1.3
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Frame = -3
Query: 466 ASVMTTTRYVRG--QPASQSYRLRLQGLLRPVRAEVAGPVGDTDDAL 332
A +MT+ Y+ G P S +YRL L+ L RAE+A D+D+ L
Sbjct: 19 ALIMTSFAYMDGVIDPPSSAYRLTLENLAEKARAEIAFVALDSDELL 65
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 470,286,749
Number of Sequences: 1393205
Number of extensions: 10108072
Number of successful extensions: 27221
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 26293
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27190
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18173652336
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)