Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002259A_C01 KMC002259A_c01
(930 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
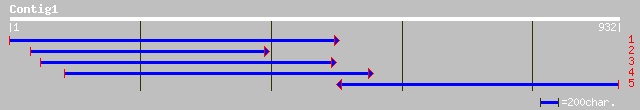
gb|AAC62625.1| rac GTPase activating protein 2 [Lotus japonicus] 495 e-139
ref|NP_182194.2| putative rac GTPase activating protein; protein... 204 2e-51
pir||C84906 probable rac GTPase activating protein [imported] - ... 204 2e-51
ref|NP_187756.1| putative rac GTPase activating protein; protein... 164 1e-39
dbj|BAC20714.1| similar to rac GTPase activating protein 3 [Oryz... 161 1e-38
>gb|AAC62625.1| rac GTPase activating protein 2 [Lotus japonicus]
Length = 424
Score = 495 bits (1274), Expect = e-139
Identities = 243/243 (100%), Positives = 243/243 (100%)
Frame = -1
Query: 930 LDSLTPEQVMHCNSEEDCTNLVKLLPSTEAALLDWAINLMADVVEHEQFNKMNARNIAMV 751
LDSLTPEQVMHCNSEEDCTNLVKLLPSTEAALLDWAINLMADVVEHEQFNKMNARNIAMV
Sbjct: 182 LDSLTPEQVMHCNSEEDCTNLVKLLPSTEAALLDWAINLMADVVEHEQFNKMNARNIAMV 241
Query: 750 FAPNMTQMVDPLTALIHAVQVMNFLKTLILKTLRERDESMAKARQLSSLLNSPSCKGDSH 571
FAPNMTQMVDPLTALIHAVQVMNFLKTLILKTLRERDESMAKARQLSSLLNSPSCKGDSH
Sbjct: 242 FAPNMTQMVDPLTALIHAVQVMNFLKTLILKTLRERDESMAKARQLSSLLNSPSCKGDSH 301
Query: 570 PFKDNREESSAQPVDTCATMPPDKSEFSRMEWCVDEKVWSSEEKGTGGGALESVSGGSSP 391
PFKDNREESSAQPVDTCATMPPDKSEFSRMEWCVDEKVWSSEEKGTGGGALESVSGGSSP
Sbjct: 302 PFKDNREESSAQPVDTCATMPPDKSEFSRMEWCVDEKVWSSEEKGTGGGALESVSGGSSP 361
Query: 390 SRYESGPLESRYRGIYDSEHWLRLRKGVRRLCQHPVFQLSKSTKKRADLGIVNTREGGGE 211
SRYESGPLESRYRGIYDSEHWLRLRKGVRRLCQHPVFQLSKSTKKRADLGIVNTREGGGE
Sbjct: 362 SRYESGPLESRYRGIYDSEHWLRLRKGVRRLCQHPVFQLSKSTKKRADLGIVNTREGGGE 421
Query: 210 AWA 202
AWA
Sbjct: 422 AWA 424
>ref|NP_182194.2| putative rac GTPase activating protein; protein id: At2g46710.1
[Arabidopsis thaliana] gi|26449717|dbj|BAC41982.1|
putative rac GTPase activating protein [Arabidopsis
thaliana] gi|28951019|gb|AAO63433.1| At2g46710
[Arabidopsis thaliana]
Length = 455
Score = 204 bits (519), Expect = 2e-51
Identities = 124/251 (49%), Positives = 150/251 (59%), Gaps = 8/251 (3%)
Frame = -1
Query: 930 LDSLTPEQVMHCNSEEDCTNLVKLLPSTEAALLDWAINLMADVVEHEQFNKMNARNIAMV 751
LD LTPEQVM CN+EEDC+ LV LLP E+A+LDWAI LMADVVEHEQFNKMNARN+AMV
Sbjct: 235 LDVLTPEQVMRCNTEEDCSRLVILLPPVESAILDWAIGLMADVVEHEQFNKMNARNVAMV 294
Query: 750 FAPNMTQMVDPLTALIHAVQVMNFLKTLILKTLRERDESMAKARQLSSLLNSPSCKGDSH 571
FAPNMTQM DPLTALIHAVQVMNFLKTLIL L+ER+ + AKAR L + PS + +S
Sbjct: 295 FAPNMTQMADPLTALIHAVQVMNFLKTLILMNLKERENADAKARWLKKQTSDPSEEWESQ 354
Query: 570 ------PFKDNREESSAQPVDTCATMPPDKSEFSRMEWCVDEKVWSSEEKGTGGGALESV 409
P K N V T + D +E+ W+ +++ G L++
Sbjct: 355 HSEILSPEKPNNNNPKFLRVATLCRLEADN----------EEEFWNIKKRNDHEGVLDTS 404
Query: 408 SGGSSPSRYESGPLESRYRGIYDSEHWLRLRKGVRRLCQHPVFQLSKSTKKRADLGIVNT 229
SG + GP V+RLC+HP+FQLSKSTKK + N
Sbjct: 405 SGNGN-----IGP--------------------VQRLCKHPLFQLSKSTKKAF---VSNR 436
Query: 228 REG--GGEAWA 202
EG G EAW+
Sbjct: 437 DEGRKGREAWS 447
>pir||C84906 probable rac GTPase activating protein [imported] - Arabidopsis
thaliana gi|3831445|gb|AAC69928.1| putative rac GTPase
activating protein [Arabidopsis thaliana]
Length = 301
Score = 204 bits (519), Expect = 2e-51
Identities = 124/251 (49%), Positives = 150/251 (59%), Gaps = 8/251 (3%)
Frame = -1
Query: 930 LDSLTPEQVMHCNSEEDCTNLVKLLPSTEAALLDWAINLMADVVEHEQFNKMNARNIAMV 751
LD LTPEQVM CN+EEDC+ LV LLP E+A+LDWAI LMADVVEHEQFNKMNARN+AMV
Sbjct: 81 LDVLTPEQVMRCNTEEDCSRLVILLPPVESAILDWAIGLMADVVEHEQFNKMNARNVAMV 140
Query: 750 FAPNMTQMVDPLTALIHAVQVMNFLKTLILKTLRERDESMAKARQLSSLLNSPSCKGDSH 571
FAPNMTQM DPLTALIHAVQVMNFLKTLIL L+ER+ + AKAR L + PS + +S
Sbjct: 141 FAPNMTQMADPLTALIHAVQVMNFLKTLILMNLKERENADAKARWLKKQTSDPSEEWESQ 200
Query: 570 ------PFKDNREESSAQPVDTCATMPPDKSEFSRMEWCVDEKVWSSEEKGTGGGALESV 409
P K N V T + D +E+ W+ +++ G L++
Sbjct: 201 HSEILSPEKPNNNNPKFLRVATLCRLEADN----------EEEFWNIKKRNDHEGVLDTS 250
Query: 408 SGGSSPSRYESGPLESRYRGIYDSEHWLRLRKGVRRLCQHPVFQLSKSTKKRADLGIVNT 229
SG + GP V+RLC+HP+FQLSKSTKK + N
Sbjct: 251 SGNGN-----IGP--------------------VQRLCKHPLFQLSKSTKKAF---VSNR 282
Query: 228 REG--GGEAWA 202
EG G EAW+
Sbjct: 283 DEGRKGREAWS 293
>ref|NP_187756.1| putative rac GTPase activating protein; protein id: At3g11490.1
[Arabidopsis thaliana]
gi|12322911|gb|AAG51449.1|AC008153_22 putative rac
GTPase activating protein; 62102-60058 [Arabidopsis
thaliana]
Length = 435
Score = 164 bits (416), Expect = 1e-39
Identities = 97/178 (54%), Positives = 119/178 (66%), Gaps = 10/178 (5%)
Frame = -1
Query: 930 LDSLTPEQVMHCNSEEDCTNLVKLLPSTEAALLDWAINLMADVVEHEQFNKMNARNIAMV 751
LDSL+PEQVM SE++C LV+LLPSTEA+LLDWAINLMADVVE EQ NKMNARNIAMV
Sbjct: 223 LDSLSPEQVMESESEDECVELVRLLPSTEASLLDWAINLMADVVEMEQLNKMNARNIAMV 282
Query: 750 FAPNMTQMVDPLTALIHAVQVMNFLKTLILKTLRERDESMAKARQLSSLL------NSPS 589
FAPNMTQM+DPLTAL++AVQVMNFLKTLI+KTL++R ES K S+ + S
Sbjct: 283 FAPNMTQMLDPLTALMYAVQVMNFLKTLIVKTLKDRKESRDKLVPASNPSPRDHNGDQSS 342
Query: 588 CKGDSHPFKDNREES----SAQPVDTCATMPPDKSEFSRMEWCVDEKVWSSEEKGTGG 427
+ H K N+EE+ A+ D + ++ E + VD K S +GG
Sbjct: 343 SRQLLHLMKANKEETLDNFEAEMKDKEESADEEEEECAESVELVDIKKSSLVNNSSGG 400
>dbj|BAC20714.1| similar to rac GTPase activating protein 3 [Oryza sativa (japonica
cultivar-group)]
Length = 487
Score = 161 bits (408), Expect = 1e-38
Identities = 79/120 (65%), Positives = 96/120 (79%)
Frame = -1
Query: 930 LDSLTPEQVMHCNSEEDCTNLVKLLPSTEAALLDWAINLMADVVEHEQFNKMNARNIAMV 751
LDSL+PEQV+HCN+EE+C LV+LLP T+AALL+W + MADVVE E+ NKMNARN+AMV
Sbjct: 206 LDSLSPEQVLHCNTEEECVELVRLLPPTQAALLNWVVEFMADVVEEEESNKMNARNVAMV 265
Query: 750 FAPNMTQMVDPLTALIHAVQVMNFLKTLILKTLRERDESMAKARQLSSLLNSPSCKGDSH 571
FAPNMTQM DPLTAL+HAVQVMN LKTLILKTLRER+ ++ +SS +S + H
Sbjct: 266 FAPNMTQMSDPLTALMHAVQVMNLLKTLILKTLREREHDESEYSAISSQSSSSDELDEMH 325
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 764,509,176
Number of Sequences: 1393205
Number of extensions: 16899155
Number of successful extensions: 56293
Number of sequences better than 10.0: 168
Number of HSP's better than 10.0 without gapping: 52390
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 56142
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 51582455952
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)