Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002254A_C01 KMC002254A_c01
(597 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
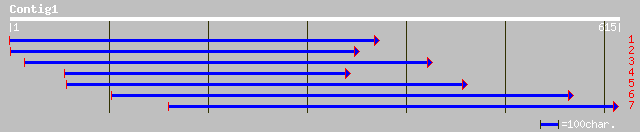
ref|NP_192993.1| F-box protein family; protein id: At4g12560.1 [... 40 0.023
gb|EAA21697.1| hypothetical protein [Plasmodium yoelii yoelii] 33 2.8
ref|NP_781876.1| chromosome segregation protein smc2 [Clostridiu... 33 3.6
ref|NP_193970.1| putative protein; protein id: At4g22390.1 [Arab... 32 6.2
ref|XP_137440.1| hypothetical protein XP_137440 [Mus musculus] 32 8.1
>ref|NP_192993.1| F-box protein family; protein id: At4g12560.1 [Arabidopsis
thaliana] gi|7487230|pir||T07648 hypothetical protein
T1P17.150 - Arabidopsis thaliana
gi|4725955|emb|CAB41726.1| putative protein [Arabidopsis
thaliana] gi|7267958|emb|CAB78299.1| putative protein
[Arabidopsis thaliana]
Length = 408
Score = 40.0 bits (92), Expect = 0.023
Identities = 18/40 (45%), Positives = 27/40 (67%)
Frame = -2
Query: 497 LGVLRGCLCISQNNKETHSFVVSQMKEYGVNESWTQLFNI 378
+GVL GCLC+ N +++ V MKEY V +SWT++F +
Sbjct: 265 IGVLDGCLCLMCNYDQSY-VDVWMMKEYNVRDSWTKVFTV 303
>gb|EAA21697.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 730
Score = 33.1 bits (74), Expect = 2.8
Identities = 13/28 (46%), Positives = 20/28 (71%)
Frame = -1
Query: 270 YNQRDNKLEDTKIANNIVGSYVRNYIES 187
Y+ DN L++TK++NNI+ S N IE+
Sbjct: 138 YHSDDNSLKETKLSNNIIASLANNSIEN 165
>ref|NP_781876.1| chromosome segregation protein smc2 [Clostridium tetani E88]
gi|28203371|gb|AAO35813.1| chromosome segregation
protein smc2 [Clostridium tetani E88]
Length = 1186
Score = 32.7 bits (73), Expect = 3.6
Identities = 19/54 (35%), Positives = 30/54 (55%)
Frame = -3
Query: 175 FLKVNKSGTF*LSPLDLIPKLVSFSSLRWDK*ILLNRDFLNIACDLINFKDWIS 14
+LK+NK G PL++I +S+ K I ++ IAC+LIN+K+ S
Sbjct: 571 YLKINKLGRATFLPLNIIKG----NSIELSKDIKNTSGYIGIACELINYKEEFS 620
>ref|NP_193970.1| putative protein; protein id: At4g22390.1 [Arabidopsis thaliana]
gi|25407584|pir||D85256 hypothetical protein AT4g22390
[imported] - Arabidopsis thaliana
gi|5738370|emb|CAB52813.1| putative protein [Arabidopsis
thaliana] gi|7269085|emb|CAB79194.1| putative protein
[Arabidopsis thaliana]
Length = 394
Score = 32.0 bits (71), Expect = 6.2
Identities = 15/40 (37%), Positives = 26/40 (64%)
Frame = -2
Query: 497 LGVLRGCLCISQNNKETHSFVVSQMKEYGVNESWTQLFNI 378
+GVL GC+C+ ++ +H V +KEY +SWT+L+ +
Sbjct: 261 IGVLDGCVCLMCYDEYSH-VDVWVLKEYEDYKSWTKLYRV 299
>ref|XP_137440.1| hypothetical protein XP_137440 [Mus musculus]
Length = 101
Score = 31.6 bits (70), Expect = 8.1
Identities = 10/20 (50%), Positives = 16/20 (80%)
Frame = -3
Query: 346 VLLCACLRMVMSCCLQSAGV 287
+L+C C+ M++ CC+ SAGV
Sbjct: 9 MLVCCCIHMLVCCCIHSAGV 28
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 495,347,684
Number of Sequences: 1393205
Number of extensions: 10229443
Number of successful extensions: 21214
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 20711
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21204
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)