Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002252A_C01 KMC002252A_c01
(1093 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
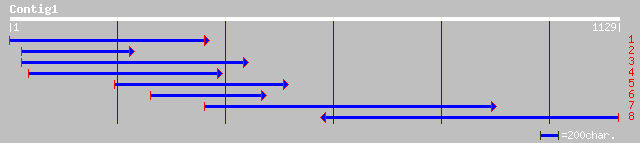
sp|Q41112|SRP_PHAVU Stress-related protein (PvSRP) gi|7488786|pi... 265 1e-69
ref|NP_187201.1| stress related protein, putative; protein id: A... 263 3e-69
sp|Q9SW70|SRP_VITRI Stress-related protein gi|5802955|gb|AAD5185... 228 1e-58
dbj|BAC15991.1| putative stress-related protein [Oryza sativa (j... 199 5e-50
ref|NP_182299.1| unknown protein; protein id: At2g47780.1 [Arabi... 195 9e-49
>sp|Q41112|SRP_PHAVU Stress-related protein (PvSRP) gi|7488786|pir||T11750 stress
related protein - kidney bean gi|1326163|gb|AAB00555.1|
stress related protein PvSRP
Length = 167
Score = 265 bits (676), Expect = 1e-69
Identities = 137/173 (79%), Positives = 149/173 (85%)
Frame = -1
Query: 925 MRFAILYSNAKERAGPLKPGVDTVEEAVKTVVGPVYDKFHQVPAEFLRYVDRKVDESVSE 746
MR AILYS AKERAGPLKPGV+TVE+AVKTVV PVYD+FH VP E L+Y DRKV E
Sbjct: 1 MRCAILYSYAKERAGPLKPGVNTVEDAVKTVVAPVYDRFHLVPVELLKYADRKV----GE 56
Query: 745 IDRHVPSNVKSVSFQARSVVSEVRRAGVVESASGLARTVYSKYEPKAEQCAVSAWRRLNQ 566
+DRHVPSNVK VS QARSVVSEVRR GV S A+TVYSKYEP AEQCAVSAWR+LNQ
Sbjct: 57 LDRHVPSNVKKVSSQARSVVSEVRRDGV----STFAKTVYSKYEPTAEQCAVSAWRKLNQ 112
Query: 565 LPLFPQVAGAVLPKAAYCSEKYNEAVVSTAQKGYRVSAYLPLVPTDKIAKVFS 407
LPLFPQVA AVLPKAAYC+EKYNE +VS+A+KGYRVSAYLPLVPT+KIAKVFS
Sbjct: 113 LPLFPQVANAVLPKAAYCTEKYNEVIVSSAEKGYRVSAYLPLVPTEKIAKVFS 165
>ref|NP_187201.1| stress related protein, putative; protein id: At3g05500.1, supported
by cDNA: 13300., supported by cDNA: gi_15450630,
supported by cDNA: gi_17380639 [Arabidopsis thaliana]
gi|14424101|sp|Q9MA63|Y350_ARATH Hypothetical protein
At3g05500 gi|7596762|gb|AAF64533.1| stress related
protein, putative [Arabidopsis thaliana]
gi|15450631|gb|AAK96587.1| AT3g05500/F22F7_5 [Arabidopsis
thaliana] gi|17380640|gb|AAL36083.1| AT3g05500/F22F7_5
[Arabidopsis thaliana] gi|21537353|gb|AAM61694.1| stress
related protein, putative [Arabidopsis thaliana]
Length = 246
Score = 263 bits (673), Expect = 3e-69
Identities = 131/230 (56%), Positives = 179/230 (76%), Gaps = 22/230 (9%)
Frame = -1
Query: 1030 SQQQLLQSQPQMVEDQKQKELKYLEFVQFATIQALMRFAILYSNAKERAGPLKPGVDTVE 851
+Q L Q + M +++K++ LKYL+FVQ A ++AL+RFA++Y+ AK+++GPLKPGV++VE
Sbjct: 3 TQTDLAQPKLDMTKEEKER-LKYLQFVQAAAVEALLRFALIYAKAKDKSGPLKPGVESVE 61
Query: 850 EAVKTVVGPVYDKFHQVPAEFLRYVDRKVDESVSEIDRHVPSNVKSVSFQ---------- 701
AVKTVVGPVY+K+H VP E L+Y+D+KVD SV+E+DR VP VK VS Q
Sbjct: 62 GAVKTVVGPVYEKYHDVPVEVLKYMDQKVDMSVTELDRRVPPVVKQVSAQAISAAQIAPI 121
Query: 700 -ARSVVSEVRRAGVVESASGLARTVYSK-----------YEPKAEQCAVSAWRRLNQLPL 557
AR++ SEVRRAGVVE+ASG+A++VYSK YEPKAEQCAVSAW++LNQLPL
Sbjct: 122 VARALASEVRRAGVVETASGMAKSVYSKYEPAAKELYANYEPKAEQCAVSAWKKLNQLPL 181
Query: 556 FPQVAGAVLPKAAYCSEKYNEAVVSTAQKGYRVSAYLPLVPTDKIAKVFS 407
FP++A +P AA+CSEKYN+ VV A+KGYRV++Y+PLVPT++I+K+F+
Sbjct: 182 FPRLAQVAVPTAAFCSEKYNDTVVKAAEKGYRVTSYMPLVPTERISKIFA 231
>sp|Q9SW70|SRP_VITRI Stress-related protein gi|5802955|gb|AAD51854.1|AF178990_1 stress
related protein [Vitis riparia]
Length = 248
Score = 228 bits (581), Expect = 1e-58
Identities = 125/233 (53%), Positives = 161/233 (68%), Gaps = 22/233 (9%)
Frame = -1
Query: 1042 SDSESQQQLLQSQPQMVEDQKQKELKYLEFVQFATIQALMRFAILYSNAKERAGPLKPGV 863
++SE++QQ P+ V + +K LKYL+FVQ A I ++ F+ LY AKE +GPLKPGV
Sbjct: 2 AESEAKQQ-----PETVHGE-EKRLKYLDFVQVAAIYVIVCFSSLYEYAKENSGPLKPGV 55
Query: 862 DTVEEAVKTVVGPVYDKFHQVPAEFLRYVDRKVDESVSEIDRHVPSNVKSVSFQ------ 701
TVE VKTV+GPVY+KF+ VP E L +VDRKV+ S+ E++RHVPS VK S Q
Sbjct: 56 QTVEGTVKTVIGPVYEKFYDVPFELLMFVDRKVEASIYELERHVPSLVKRASCQAITVAQ 115
Query: 700 -----ARSVVSEVRRAGVVESASGLARTVY-----------SKYEPKAEQCAVSAWRRLN 569
A +V SEV+R GVV++A + + VY SKYEP AEQ AVSAWR LN
Sbjct: 116 KAPELALAVASEVQRPGVVDTAKNITKNVYSKCEPTAKELCSKYEPVAEQYAVSAWRSLN 175
Query: 568 QLPLFPQVAGAVLPKAAYCSEKYNEAVVSTAQKGYRVSAYLPLVPTDKIAKVF 410
+LPLFPQVA V+P AAY SEKYN++V TA++GY V+ YLPL+PT++IAKVF
Sbjct: 176 RLPLFPQVAQVVVPTAAYWSEKYNQSVSYTAERGYTVALYLPLIPTERIAKVF 228
>dbj|BAC15991.1| putative stress-related protein [Oryza sativa (japonica
cultivar-group)]
Length = 255
Score = 199 bits (507), Expect = 5e-50
Identities = 106/224 (47%), Positives = 143/224 (63%), Gaps = 29/224 (12%)
Frame = -1
Query: 994 VEDQKQKELKYLEFVQFATIQALMRFAILYSNAKERAGPLKPGVDTVEEAVKTVVGPVYD 815
VE+ ++ +L+YLEFVQ A QA + A LY+ AK+ AGPL+PGVD VE AVK VVGPVY
Sbjct: 14 VEEVRRPKLRYLEFVQVAAAQATICLAGLYALAKDHAGPLRPGVDAVESAVKGVVGPVYG 73
Query: 814 KFHQVPAEFLRYVDRKVDESVSEIDRHVPSNVKSVSFQ-----------ARSVVSEVRRA 668
+FH VP + L +VDRKVD++V E+DRH+P +K+ S + AR + +EV+++
Sbjct: 74 RFHGVPLDVLAFVDRKVDDTVQELDRHLPPTLKAASAKACAVARGVPDVARELTAEVQQS 133
Query: 667 GVVESA------------------SGLARTVYSKYEPKAEQCAVSAWRRLNQLPLFPQVA 542
GV +A A+ +Y +YEP AE AVS WR LN LPLFPQVA
Sbjct: 134 GVTGAARVAYAKVEPVAKGVYGRIQPAAKDLYVRYEPAAEHLAVSTWRSLNNLPLFPQVA 193
Query: 541 GAVLPKAAYCSEKYNEAVVSTAQKGYRVSAYLPLVPTDKIAKVF 410
+P AAY +EKYN+ + + A KGY + YLP +PT++IAKVF
Sbjct: 194 QIAVPTAAYWAEKYNKVIAAAADKGYTGAQYLPAIPTERIAKVF 237
>ref|NP_182299.1| unknown protein; protein id: At2g47780.1 [Arabidopsis thaliana]
gi|14424099|sp|O82246|Y278_ARATH Hypothetical protein
At2g47780 gi|25409061|pir||D84919 hypothetical protein
At2g47780 [imported] - Arabidopsis thaliana
gi|3738291|gb|AAC63633.1| unknown protein [Arabidopsis
thaliana] gi|23306436|gb|AAN17445.1| unknown protein
[Arabidopsis thaliana] gi|25084024|gb|AAN72157.1|
unknown protein [Arabidopsis thaliana]
Length = 235
Score = 195 bits (496), Expect = 9e-49
Identities = 95/195 (48%), Positives = 135/195 (68%)
Frame = -1
Query: 997 MVEDQKQKELKYLEFVQFATIQALMRFAILYSNAKERAGPLKPGVDTVEEAVKTVVGPVY 818
+VED + +LK+LEF+Q A + F+ LY AK+ AGPLK GV+ +E+ V+TV+ P+Y
Sbjct: 32 VVEDDDEMKLKHLEFIQVAAVYFAACFSTLYELAKDNAGPLKLGVENIEDCVRTVLAPLY 91
Query: 817 DKFHQVPAEFLRYVDRKVDESVSEIDRHVPSNVKSVSFQARSVVSEVRRAGVVESASGLA 638
+KFH VP + L +VDRKVD+ +++ +VPS VK S QA +V +EV+R GVV++ +A
Sbjct: 92 EKFHDVPFKLLLFVDRKVDDVFFDVETYVPSLVKQASSQALTVATEVQRTGVVDTTKSIA 151
Query: 637 RTVYSKYEPKAEQCAVSAWRRLNQLPLFPQVAGAVLPKAAYCSEKYNEAVVSTAQKGYRV 458
R+V KYEP AE A + WR LNQLPLFP+VA V+P A Y SEKYN+AV + Y
Sbjct: 152 RSVRDKYEPAAEYYAATLWRLLNQLPLFPEVAHLVIPTAFYWSEKYNDAVRYVGDRDYFG 211
Query: 457 SAYLPLVPTDKIAKV 413
+ YLP++P +KI+ +
Sbjct: 212 AEYLPMIPIEKISDI 226
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,005,582,439
Number of Sequences: 1393205
Number of extensions: 24579363
Number of successful extensions: 107546
Number of sequences better than 10.0: 566
Number of HSP's better than 10.0 without gapping: 84011
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 103648
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 65340192036
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)