Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002251A_C01 KMC002251A_c01
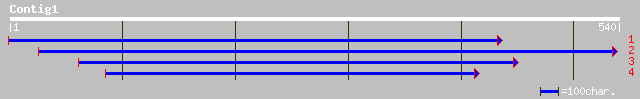
(540 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_182092.1| putative PCF2-like DNA binding protein; protein... 49 4e-05
ref|XP_209582.1| hypothetical protein XP_209582 [Homo sapiens] 42 0.004
gb|AAO52512.1| similar to Streptococcus pneumoniae. Cell wall su... 40 0.014
ref|NP_181536.1| En/Spm-like transposon protein; protein id: At2... 40 0.018
gb|EAA33270.1| hypothetical protein [Neurospora crassa] 40 0.024
>ref|NP_182092.1| putative PCF2-like DNA binding protein; protein id: At2g45680.1,
supported by cDNA: gi_13877694, supported by cDNA:
gi_15810350, supported by cDNA: gi_21281084 [Arabidopsis
thaliana] gi|7485632|pir||T00881 probable PCF2-like DNA
binding protein [imported] - Arabidopsis thaliana
gi|2979559|gb|AAC06168.1| putative PCF2-like DNA binding
protein [Arabidopsis thaliana]
gi|13877695|gb|AAK43925.1|AF370606_1 putative PCF2-like
DNA binding protein [Arabidopsis thaliana]
gi|15810351|gb|AAL07063.1| putative PCF2 DNA binding
protein [Arabidopsis thaliana]
gi|21281085|gb|AAM45029.1| putative PCF2 DNA binding
protein [Arabidopsis thaliana]
Length = 356
Score = 48.9 bits (115), Expect = 4e-05
Identities = 27/50 (54%), Positives = 37/50 (74%), Gaps = 11/50 (22%)
Frame = -1
Query: 537 TVMAPSTTT----------TTTTTHTLRDFSLEIYDRQEL-QFMSRSSSK 421
+VMAPS+++ TTTTHTLRDFSLEIY++QEL QFMS ++++
Sbjct: 303 SVMAPSSSSGVTTGSSSSIATTTTHTLRDFSLEIYEKQELHQFMSTTTAR 352
>ref|XP_209582.1| hypothetical protein XP_209582 [Homo sapiens]
Length = 252
Score = 42.4 bits (98), Expect = 0.004
Identities = 33/125 (26%), Positives = 60/125 (47%)
Frame = +3
Query: 117 ILINVIIIRKITSSGISSTSTPSCTVHGASTTSGTPPSPPLFVLTHTPHTQVVPTLFNPT 296
IL + I+ I SS SS+S S + ++S + SP T +P + + +P+
Sbjct: 45 ILSSSILSSSIPSSSSSSSSPSSSPSSSSPSSSHSSSSPSSSSSTSSPSSSSSSSSSSPS 104
Query: 297 HVRTCRSLFSFSNDVVNGGTQNKPEPNSNVSSSSSSSSSLNASRKNAT*TEALACRKSPR 476
+ S S S + + + P +S+ SSSSSSSS + S +++ + + + S
Sbjct: 105 SSNSSSSSSSSSPSSSSSSSSSSPSSSSSSPSSSSSSSSSSPSSSSSSPSSSSSSSSSSS 164
Query: 477 RNPSA 491
+PS+
Sbjct: 165 SSPSS 169
Score = 39.3 bits (90), Expect = 0.031
Identities = 34/121 (28%), Positives = 61/121 (50%), Gaps = 1/121 (0%)
Frame = +3
Query: 141 RKITSSGISSTSTPSCTVHGASTTSGTPPSPPLFVLTHTPHTQVVPTLFNPTHVRTCRSL 320
R I SS I S+S PS +S++S +P S P + H+ P+ + T + S
Sbjct: 43 RSILSSSILSSSIPS-----SSSSSSSPSSSPSSSSPSSSHSSSSPSSSSSTSSPSSSSS 97
Query: 321 FSFSNDVVNGGTQNKPEPNSNVSSSSSSSS-SLNASRKNAT*TEALACRKSPRRNPSACA 497
S S+ + + + + + SSSSSSSS S ++S +++ + + + S +PS+ +
Sbjct: 98 SSSSSPSSSNSSSSSSSSSPSSSSSSSSSSPSSSSSSPSSSSSSSSSSPSSSSSSPSSSS 157
Query: 498 S 500
S
Sbjct: 158 S 158
Score = 35.8 bits (81), Expect = 0.34
Identities = 30/117 (25%), Positives = 57/117 (48%)
Frame = +3
Query: 150 TSSGISSTSTPSCTVHGASTTSGTPPSPPLFVLTHTPHTQVVPTLFNPTHVRTCRSLFSF 329
+SS SS+S+PS + +S++S +P S + + P+ S
Sbjct: 93 SSSSSSSSSSPSSSNSSSSSSSSSPSSSSSSSSSSPSSSSSSPS--------------SS 138
Query: 330 SNDVVNGGTQNKPEPNSNVSSSSSSSSSLNASRKNAT*TEALACRKSPRRNPSACAS 500
S+ + + + P+S+ SSSSSSSSS ++S +++ + + SP + S+ +S
Sbjct: 139 SSSSSSSPSSSSSSPSSSSSSSSSSSSSPSSSSPSSSGSSPSSSNSSPSSSSSSPSS 195
Score = 32.3 bits (72), Expect = 3.8
Identities = 26/107 (24%), Positives = 53/107 (49%), Gaps = 3/107 (2%)
Frame = +3
Query: 150 TSSGISSTSTPSCTVHGASTTSGTPPSPPLFVLTHTPHTQVVPTLFNPTHVRTCRSLFS- 326
+SS SS+S+PS + +S++ + S P + + + P+ +P+ + S +
Sbjct: 125 SSSPSSSSSSPSSSSSSSSSSPSSSSSSPSSSSSSSSSSSSSPSSSSPSSSGSSPSSSNS 184
Query: 327 --FSNDVVNGGTQNKPEPNSNVSSSSSSSSSLNASRKNAT*TEALAC 461
S+ + + P P S+ SSSSSS+S +S ++ + + +C
Sbjct: 185 SPSSSSSSPSSSSSSPSPRSSSPSSSSSSTSSPSSSSPSSSSPSSSC 231
Score = 31.2 bits (69), Expect = 8.4
Identities = 28/119 (23%), Positives = 58/119 (48%), Gaps = 3/119 (2%)
Frame = +3
Query: 153 SSGISSTSTPSCTVHGASTT---SGTPPSPPLFVLTHTPHTQVVPTLFNPTHVRTCRSLF 323
SS SS+S+PS + +S++ S + PS + +P + + + + S
Sbjct: 108 SSSSSSSSSPSSSSSSSSSSPSSSSSSPSSSSSSSSSSPSSSSSSPSSSSSSSSSSSSSP 167
Query: 324 SFSNDVVNGGTQNKPEPNSNVSSSSSSSSSLNASRKNAT*TEALACRKSPRRNPSACAS 500
S S+ +G + + + + SSSS SSSS + S ++++ + + + SP + + +S
Sbjct: 168 SSSSPSSSGSSPSSSNSSPSSSSSSPSSSSSSPSPRSSSPSSSSSSTSSPSSSSPSSSS 226
>gb|AAO52512.1| similar to Streptococcus pneumoniae. Cell wall surface anchor family
protein [Dictyostelium discoideum]
Length = 1806
Score = 40.4 bits (93), Expect = 0.014
Identities = 33/95 (34%), Positives = 49/95 (50%), Gaps = 3/95 (3%)
Frame = +3
Query: 165 SSTSTPSCTVHGASTT-SGTPPSPPLFVLTHTPHTQVVPT--LFNPTHVRTCRSLFSFSN 335
S+++TPS + G+S++ + T PS LF T + T P+ LF T T + F
Sbjct: 1379 STSTTPSTGLFGSSSSIATTTPSTGLFGSTSSSTTNTAPSTGLFGSTTTSTTATPF---- 1434
Query: 336 DVVNGGTQNKPEPNSNVSSSSSSSSSLNASRKNAT 440
G + + P SSSSS+SSSL++S AT
Sbjct: 1435 ----GSSSSTPSTGLFGSSSSSTSSSLSSSSTTAT 1465
Score = 34.3 bits (77), Expect = 0.99
Identities = 31/99 (31%), Positives = 48/99 (48%)
Frame = +3
Query: 150 TSSGISSTSTPSCTVHGASTTSGTPPSPPLFVLTHTPHTQVVPTLFNPTHVRTCRSLFSF 329
T++ ++T+TPS ++ G++TT T S LT TP T + + T LF
Sbjct: 1219 TTTTAATTATPSTSLFGSTTTPSTSSSTSS--LTTTPSTGLFGASSSTT---PSTGLF-- 1271
Query: 330 SNDVVNGGTQNKPEPNSNVSSSSSSSSSLNASRKNAT*T 446
G+ P +SSSSSSSS+++S +T T
Sbjct: 1272 -------GSATTPSTGLFGASSSSSSSSISSSSTTSTTT 1303
Score = 32.0 bits (71), Expect = 4.9
Identities = 33/105 (31%), Positives = 47/105 (44%), Gaps = 6/105 (5%)
Frame = +3
Query: 144 KITSSGISSTSTPSCTVHGASTTSGTPPSPPLFVLTHTPHTQVVPTLF--NPT-HVRTCR 314
KI ++T+T + ++ G++TTSG L P T +LF NP +
Sbjct: 1032 KIDDKPSTTTTTTTTSLFGSTTTSG---------LFSNPSTTSTGSLFSSNPLGSTASTS 1082
Query: 315 SLFSFSNDVVNGGTQNKPEPN---SNVSSSSSSSSSLNASRKNAT 440
SLF S T P + S SSSSSSSS ++S + T
Sbjct: 1083 SLFGASTLASTATTTATPTTSLFGSTTPSSSSSSSSSSSSTTSTT 1127
>ref|NP_181536.1| En/Spm-like transposon protein; protein id: At2g40070.1
[Arabidopsis thaliana] gi|25408663|pir||H84824
En/Spm-like transposon protein [imported] - Arabidopsis
thaliana gi|2088658|gb|AAB95292.1| En/Spm-like
transposon protein [Arabidopsis thaliana]
Length = 510
Score = 40.0 bits (92), Expect = 0.018
Identities = 33/114 (28%), Positives = 53/114 (45%), Gaps = 1/114 (0%)
Frame = +3
Query: 162 ISSTSTPSCTVHGASTTSGTPPSPPLFVLTHTPHTQVVPTLFNPTHVRTCRSLFSFSNDV 341
+SS + PS T + ST S T P+ T +++ PT PT T RS S +
Sbjct: 187 VSSATRPSLT-NSRSTVSATTKPTPMSRSTSLSSSRLTPTASKPT-TSTARSAGSVTRST 244
Query: 342 VNGGTQNK-PEPNSNVSSSSSSSSSLNASRKNAT*TEALACRKSPRRNPSACAS 500
+ T++ P ++ S S++ SS SR ++ ++ +P R P A AS
Sbjct: 245 PSTTTKSAGPSRSTTPLSRSTARSSTPTSRPTLPPSKTISRSSTPTRRPIASAS 298
>gb|EAA33270.1| hypothetical protein [Neurospora crassa]
Length = 318
Score = 39.7 bits (91), Expect = 0.024
Identities = 26/61 (42%), Positives = 31/61 (50%), Gaps = 11/61 (18%)
Frame = -3
Query: 535 GHGSE---HHHHHHHDAHAEGF------LLG--DLRQARASVHVAFFLEALREEEEEEEE 389
GHG++ HHHHHHH H+ F LG L + + V LREEEE EEE
Sbjct: 207 GHGNDGGPHHHHHHHLQHSSTFGQQRERALGGQQLNGSGSDVQSKEGEALLREEEEGEEE 266
Query: 388 E 386
E
Sbjct: 267 E 267
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 503,469,863
Number of Sequences: 1393205
Number of extensions: 12318670
Number of successful extensions: 181656
Number of sequences better than 10.0: 623
Number of HSP's better than 10.0 without gapping: 85675
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 148624
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)