Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
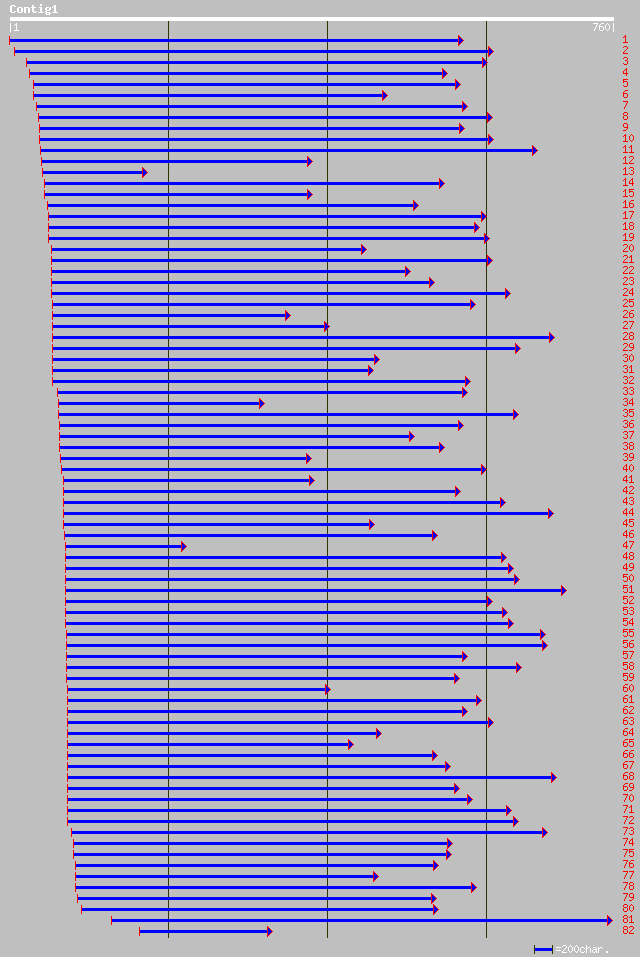
Query= KMC002238A_C01 KMC002238A_c01
(710 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|EAA00692.1| ebiP8665 [Anopheles gambiae str. PEST] 40 0.042
dbj|BAA85438.1| similar to RING-H2 finger protein RHA1a (AF07868... 36 0.46
gb|ZP_00004281.1| hypothetical protein [Rhodobacter sphaeroides] 35 0.79
gb|EAA36420.1| hypothetical protein [Neurospora crassa] 35 1.3
gb|AAK83835.1| glycine-rich protein GRP17 [Arabidopsis thaliana] 34 1.8
>gb|EAA00692.1| ebiP8665 [Anopheles gambiae str. PEST]
Length = 2010
Score = 39.7 bits (91), Expect = 0.042
Identities = 20/59 (33%), Positives = 33/59 (55%)
Frame = +1
Query: 466 EDSKHQQQS*EQSRRWRSRTGSSGGDNGGSGETQRQRRL*KGGSGGHRAKETVA*GKIK 642
++ + +++ E+ R +S+ G SGG GG G + + KGG GG+ K+ V KIK
Sbjct: 263 DEDEEEEEEVERERTRKSKKGKSGGGGGGGGSSSSNK---KGGGGGNSRKQKVPTLKIK 318
>dbj|BAA85438.1| similar to RING-H2 finger protein RHA1a (AF078683) [Oryza sativa
(japonica cultivar-group)] gi|25553533|dbj|BAC24809.1|
OSJNBa0004I20.4 [Oryza sativa (japonica cultivar-group)]
Length = 224
Score = 36.2 bits (82), Expect = 0.46
Identities = 20/50 (40%), Positives = 26/50 (52%)
Frame = +2
Query: 11 FFFFFDFNFFFFWSMVFGNRYPKGLSSGAYISVRINLKRALHKCTYIRRL 160
FFFFF F FFFF+ + + + G G Y V L L T++RRL
Sbjct: 48 FFFFFFFFFFFFFILSWTTKNIMGFPVGGYSGVPRLLLHLLFLLTHLRRL 97
>gb|ZP_00004281.1| hypothetical protein [Rhodobacter sphaeroides]
Length = 305
Score = 35.4 bits (80), Expect = 0.79
Identities = 24/81 (29%), Positives = 38/81 (46%), Gaps = 3/81 (3%)
Frame = +2
Query: 473 PNTSSKAENKVGDGGVGL-VAAVVITAAAARPSGRDGCRRADLVVTEQRRQLHE--GRSS 643
P +++ ++ G G GL AA ++ A A PSG RR ++ R L G +
Sbjct: 12 PVPAARGADERGADGHGLPAAAALLRACGAEPSGCPALRRQGQLLRGAARALSRAAGHRA 71
Query: 644 GHGCKRRKKRLQSKWQGRGER 706
H +R + RL + GR +R
Sbjct: 72 AHAAQRARGRLHRRGAGRADR 92
>gb|EAA36420.1| hypothetical protein [Neurospora crassa]
Length = 1171
Score = 34.7 bits (78), Expect = 1.3
Identities = 24/63 (38%), Positives = 32/63 (50%), Gaps = 6/63 (9%)
Frame = +1
Query: 442 IGGKRKTFEDSK------HQQQS*EQSRRWRSRTGSSGGDNGGSGETQRQRRL*KGGSGG 603
+GG R ++ + QQQ+ Q S GSSGG GG G+ Q+Q+ GG GG
Sbjct: 1097 LGGARSAYDYGQGWYGAGQQQQNGNQGG---SGGGSSGG-GGGHGQQQQQQHAVHGGHGG 1152
Query: 604 HRA 612
H A
Sbjct: 1153 HHA 1155
>gb|AAK83835.1| glycine-rich protein GRP17 [Arabidopsis thaliana]
Length = 543
Score = 34.3 bits (77), Expect = 1.8
Identities = 32/108 (29%), Positives = 45/108 (41%)
Frame = +1
Query: 319 AALPNSNSSDSAGGCKTCS*K*SSKSRLQQKANGVNMIDLRIGGKRKTFEDSKHQQQS*E 498
A LP NS AGG ++ K +KS+ KA M+ + GGK+ K
Sbjct: 154 AGLP-PNSGAGAGGAQSLIKKSKAKSKGGLKAWCKKMLKSKFGGKKGKSGGGKS------ 206
Query: 499 QSRRWRSRTGSSGGDNGGSGETQRQRRL*KGGSGGHRAKETVA*GKIK 642
++ + G S G+ G S +R G SGG K GK+K
Sbjct: 207 ---KFGGKGGKSEGEEGMSSGDERMSGSEGGMSGGEGGKSKSGKGKLK 251
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 619,954,382
Number of Sequences: 1393205
Number of extensions: 14088519
Number of successful extensions: 70877
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 55207
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 64845
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32654539052
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)