Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002231A_C01 KMC002231A_c01
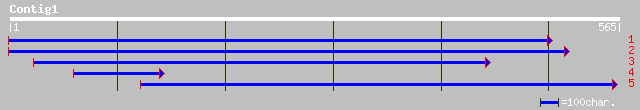
(565 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_190370.1| putative protein; protein id: At3g47860.1, supp... 205 3e-52
gb|AAB35919.1| apolipoprotein D; apoD [Homo sapiens] 39 0.044
gb|AAB32200.1| apolipoprotein D, apoD [human, plasma, Peptide, 2... 39 0.044
ref|NP_001638.1| apolipoprotein D precursor [Homo sapiens] gi|11... 39 0.044
sp|P51910|APOD_MOUSE Apolipoprotein D precursor (ApoD) gi|847651... 39 0.058
>ref|NP_190370.1| putative protein; protein id: At3g47860.1, supported by cDNA:
gi_14334983, supported by cDNA: gi_14994280 [Arabidopsis
thaliana] gi|7487370|pir||T07725 hypothetical protein
T23J7.190 - Arabidopsis thaliana
gi|4741203|emb|CAB41869.1| putative protein [Arabidopsis
thaliana] gi|14334984|gb|AAK59669.1| unknown protein
[Arabidopsis thaliana] gi|14994281|gb|AAK73275.1|
putative protein [Arabidopsis thaliana]
gi|27754734|gb|AAO22810.1| unknown protein [Arabidopsis
thaliana]
Length = 353
Score = 205 bits (522), Expect = 3e-52
Identities = 94/122 (77%), Positives = 109/122 (89%)
Frame = -3
Query: 563 PFIPKLPYDVIATDYDNFALVSGAKDRGFVQIYSRTPNPGTEFIERNKAYLENFGYDPSK 384
PFIPKLPYDVIATDYDN+ALVSGAKD+GFVQ+YSRTPNPG EFI + K YL FGYDP K
Sbjct: 232 PFIPKLPYDVIATDYDNYALVSGAKDKGFVQVYSRTPNPGPEFIAKYKNYLAQFGYDPEK 291
Query: 383 IKDTPQDCEVSDSKLSAMMSMSGMQQALTNQFPDIGLKAPIEFNPFTSVFDTFKKLVELY 204
IKDTPQDCEV+D++L+AMMSM GM+Q LTNQFPD+GL+ ++F+PFTSVF+T KKLV LY
Sbjct: 292 IKDTPQDCEVTDAELAAMMSMPGMEQTLTNQFPDLGLRKSVQFDPFTSVFETLKKLVPLY 351
Query: 203 FK 198
FK
Sbjct: 352 FK 353
>gb|AAB35919.1| apolipoprotein D; apoD [Homo sapiens]
Length = 98
Score = 38.9 bits (89), Expect = 0.044
Identities = 24/71 (33%), Positives = 34/71 (47%), Gaps = 6/71 (8%)
Frame = -3
Query: 560 FIPKLPYDVIATDYDNFALVSGAK------DRGFVQIYSRTPNPGTEFIERNKAYLENFG 399
F+P PY ++ATDY+N+ALV F I +R PN E ++ K L +
Sbjct: 21 FMPSAPYWILATDYENYALVYSCTCIIQLFHVDFAWILARNPNLPPETVDSLKNILTSNN 80
Query: 398 YDPSKIKDTPQ 366
D K+ T Q
Sbjct: 81 IDVKKMTVTDQ 91
>gb|AAB32200.1| apolipoprotein D, apoD [human, plasma, Peptide, 246 aa]
Length = 246
Score = 38.9 bits (89), Expect = 0.044
Identities = 24/71 (33%), Positives = 34/71 (47%), Gaps = 6/71 (8%)
Frame = -3
Query: 560 FIPKLPYDVIATDYDNFALVSGAK------DRGFVQIYSRTPNPGTEFIERNKAYLENFG 399
F+P PY ++ATDY+N+ALV F I +R PN E ++ K L +
Sbjct: 92 FMPSAPYWILATDYENYALVYSCTCIIQLFHVDFAWILARNPNLPPETVDSLKNILTSNN 151
Query: 398 YDPSKIKDTPQ 366
D K+ T Q
Sbjct: 152 IDVKKMTVTDQ 162
>ref|NP_001638.1| apolipoprotein D precursor [Homo sapiens]
gi|114034|sp|P05090|APOD_HUMAN Apolipoprotein D
precursor (Apo-D) (ApoD) gi|72088|pir||LPHUD
apolipoprotein D precursor [validated] - human
gi|178841|gb|AAB59517.1| apolipoprotein D precursor
gi|178847|gb|AAA51764.1| apolipoprotein D precursor
gi|13938509|gb|AAH07402.1|AAH07402 apolipoprotein D
[Homo sapiens]
Length = 189
Score = 38.9 bits (89), Expect = 0.044
Identities = 24/71 (33%), Positives = 34/71 (47%), Gaps = 6/71 (8%)
Frame = -3
Query: 560 FIPKLPYDVIATDYDNFALVSGAK------DRGFVQIYSRTPNPGTEFIERNKAYLENFG 399
F+P PY ++ATDY+N+ALV F I +R PN E ++ K L +
Sbjct: 112 FMPSAPYWILATDYENYALVYSCTCIIQLFHVDFAWILARNPNLPPETVDSLKNILTSNN 171
Query: 398 YDPSKIKDTPQ 366
D K+ T Q
Sbjct: 172 IDVKKMTVTDQ 182
>sp|P51910|APOD_MOUSE Apolipoprotein D precursor (ApoD) gi|847651|gb|AAA67892.1|
apolipoprotein D [Mus musculus]
Length = 189
Score = 38.5 bits (88), Expect = 0.058
Identities = 26/72 (36%), Positives = 33/72 (45%), Gaps = 6/72 (8%)
Frame = -3
Query: 563 PFIPKLPYDVIATDYDNFALVSGAK------DRGFVQIYSRTPNPGTEFIERNKAYLENF 402
P +P PY ++ATDY+N+ALV FV I R P E I K L +
Sbjct: 111 PLMPPAPYWILATDYENYALVYSCTTFFWLFHVDFVWILGRNPYLPPETITYLKDILTSN 170
Query: 401 GYDPSKIKDTPQ 366
G D K+ T Q
Sbjct: 171 GIDIEKMTTTDQ 182
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 432,596,345
Number of Sequences: 1393205
Number of extensions: 8473612
Number of successful extensions: 20122
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 19695
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20118
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20382500157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)