Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002227A_C01 KMC002227A_c01
(677 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
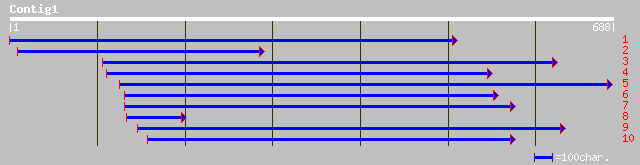
ref|NP_173107.1| arginine/serine-rich protein, putative; protein... 178 5e-44
gb|AAK76509.1| putative arginine/serine-rich protein [Arabidopsi... 176 3e-43
dbj|BAB89002.1| P0401G10.10 [Oryza sativa (japonica cultivar-gro... 86 4e-16
ref|NP_492875.2| pre-mRNA splicing SR protein related RSR-1 (68.... 83 4e-15
ref|NP_058079.1| Ser/Arg-related nuclear matrix protein; plenty-... 74 2e-12
>ref|NP_173107.1| arginine/serine-rich protein, putative; protein id: At1g16610.1,
supported by cDNA: gi_15027956, supported by cDNA:
gi_6601501 [Arabidopsis thaliana]
gi|25402809|pir||C86301 arginine/serine-rich protein
[imported] - Arabidopsis thaliana
gi|6601502|gb|AAF19004.1|AF151366_1 arginine/serine-rich
protein [Arabidopsis thaliana]
gi|9989058|gb|AAG10821.1|AC011808_9 arginine/serine-rich
protein [Arabidopsis thaliana]
Length = 414
Score = 178 bits (452), Expect = 5e-44
Identities = 95/122 (77%), Positives = 104/122 (84%), Gaps = 2/122 (1%)
Frame = -2
Query: 673 SPRRLRGSPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGRRRSRSPIRRPARSNSRSFSP 494
SPRR+RGSP RRRSP P RRRSPPPRR RSPPRRSPI RRRSRSPIRRP RS S S SP
Sbjct: 294 SPRRIRGSPVRRRSPLPL-RRRSPPPRRLRSPPRRSPI-RRRSRSPIRRPGRSRSSSISP 351
Query: 493 RRGRPPV-RRGRSSSFSDSPSPRKVSRR-SRSRSPRRPLRGGRASSNSSSSSSPPPPPPP 320
R+GR P RRGRSSS+S SPSPR++ R+ SRSRSP+RPLRG R+SSNSSSSSSPPPPPPP
Sbjct: 352 RKGRGPAGRRGRSSSYSSSPSPRRIPRKISRSRSPKRPLRGKRSSSNSSSSSSPPPPPPP 411
Query: 319 AR 314
+
Sbjct: 412 RK 413
Score = 45.8 bits (107), Expect = 5e-04
Identities = 44/128 (34%), Positives = 52/128 (40%), Gaps = 19/128 (14%)
Frame = -2
Query: 625 PPPRRRSPPPRRARSPPRRSPI-------------GRRRSR--SPIRRPARS-NSRSFSP 494
PP ++ S PP+ + P+R G RR R SP R+ S RS P
Sbjct: 176 PPRQKVSSPPKPVSAAPKRDAPKSDNAAADAEKDGGPRRPRETSPQRKTGLSPRRRSPLP 235
Query: 493 RRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPP---PPP 323
RRG P RR DSP R+ R R P R + S S SSPPP P
Sbjct: 236 RRGLSPRRRS-----PDSPHRRRPGSPIRRRGDTPPRRRPASPSRGRSPSSPPPRRYRSP 290
Query: 322 PARKP*RI 299
P P RI
Sbjct: 291 PRGSPRRI 298
Score = 44.3 bits (103), Expect = 0.002
Identities = 42/97 (43%), Positives = 43/97 (44%), Gaps = 5/97 (5%)
Frame = -2
Query: 601 PPRRARSPPRRSPIGRRRSRSPIRRPARSNSRSFSPRRGRPPVRRGRSSSFSDSPSPRKV 422
P R RSP R SRS R S SRS S R R RS S S SPS R V
Sbjct: 4 PSRGRRSPSVSGSSSRSSSRS---RSGSSPSRSISRSRSR-----SRSLSSSSSPS-RSV 54
Query: 421 SRRSRS-----RSPRRPLRGGRASSNSSSSSSPPPPP 326
S SRS +SP P R GR SPPPPP
Sbjct: 55 SSGSRSPPRRGKSPAGPARRGR---------SPPPPP 82
>gb|AAK76509.1| putative arginine/serine-rich protein [Arabidopsis thaliana]
Length = 414
Score = 176 bits (445), Expect = 3e-43
Identities = 94/122 (77%), Positives = 103/122 (84%), Gaps = 2/122 (1%)
Frame = -2
Query: 673 SPRRLRGSPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGRRRSRSPIRRPARSNSRSFSP 494
SPRR+RGSP RRRSP P RRRSPPPRR RSPPRRSPI RRRSRSPIRRP RS S S SP
Sbjct: 294 SPRRIRGSPVRRRSPLPL-RRRSPPPRRLRSPPRRSPI-RRRSRSPIRRPGRSRSSSISP 351
Query: 493 RRGRPPVRR-GRSSSFSDSPSPRKVSRR-SRSRSPRRPLRGGRASSNSSSSSSPPPPPPP 320
R+GR P R GRSSS+S SPSPR++ R+ SRSRSP+RPLRG R+SSNSSSSSSPPPPPPP
Sbjct: 352 RKGRGPAGRPGRSSSYSSSPSPRRIPRKISRSRSPKRPLRGKRSSSNSSSSSSPPPPPPP 411
Query: 319 AR 314
+
Sbjct: 412 RK 413
Score = 45.8 bits (107), Expect = 5e-04
Identities = 44/128 (34%), Positives = 52/128 (40%), Gaps = 19/128 (14%)
Frame = -2
Query: 625 PPPRRRSPPPRRARSPPRRSPI-------------GRRRSR--SPIRRPARS-NSRSFSP 494
PP ++ S PP+ + P+R G RR R SP R+ S RS P
Sbjct: 176 PPRQKVSSPPKPVSAAPKRDAPKSDNAAADAEKDGGPRRPRETSPQRKTGLSPRRRSPLP 235
Query: 493 RRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPP---PPP 323
RRG P RR DSP R+ R R P R + S S SSPPP P
Sbjct: 236 RRGLSPRRRS-----PDSPHRRRPGSPIRRRGDTPPRRRPASPSRGRSPSSPPPRRYRSP 290
Query: 322 PARKP*RI 299
P P RI
Sbjct: 291 PRGSPRRI 298
Score = 44.3 bits (103), Expect = 0.002
Identities = 42/97 (43%), Positives = 43/97 (44%), Gaps = 5/97 (5%)
Frame = -2
Query: 601 PPRRARSPPRRSPIGRRRSRSPIRRPARSNSRSFSPRRGRPPVRRGRSSSFSDSPSPRKV 422
P R RSP R SRS R S SRS S R R RS S S SPS R V
Sbjct: 4 PSRGRRSPSVSGSSSRSSSRS---RSGSSPSRSISRSRSR-----SRSLSSSSSPS-RSV 54
Query: 421 SRRSRS-----RSPRRPLRGGRASSNSSSSSSPPPPP 326
S SRS +SP P R GR SPPPPP
Sbjct: 55 SSGSRSPPRRGKSPAGPARRGR---------SPPPPP 82
>dbj|BAB89002.1| P0401G10.10 [Oryza sativa (japonica cultivar-group)]
Length = 380
Score = 86.3 bits (212), Expect = 4e-16
Identities = 64/120 (53%), Positives = 67/120 (55%), Gaps = 8/120 (6%)
Frame = -2
Query: 670 PRRLRGSPGRRRSPPPPPRRRSPPPRRAR---SPPRRSPIGRRRSRSPIRRPARSNSRSF 500
PRR GSP RRRSP PPPRRR P RR R SP RRSP G R RSP P R S
Sbjct: 185 PRRRPGSPVRRRSPSPPPRRRRSPMRRLRGSPSPRRRSP-GPIRRRSPPPPPRRPRS--- 240
Query: 499 SPRRGRPPVRRGRSSSFSDSPSPRKVSR-RSRSRSPRR----PLRGGRASSNSSSSSSPP 335
P R PP RR S SP PR+ RSRS SPR PLR GR+ S+ S S SPP
Sbjct: 241 PPGRRLPPPRR-----HSRSPPPRRPPHSRSRSISPRTRRGPPLRRGRSDSSYSRSPSPP 295
Score = 84.0 bits (206), Expect = 2e-15
Identities = 52/84 (61%), Positives = 54/84 (63%), Gaps = 3/84 (3%)
Frame = -2
Query: 673 SPRRLRGSPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGRRRSRS-PIRRPARSNSRSFS 497
SPRR P RRRSPPPPPRR PP R PP RR SRS P RRP S SRS S
Sbjct: 217 SPRRRSPGPIRRRSPPPPPRRPRSPPGRRLPPP------RRHSRSPPPRRPPHSRSRSIS 270
Query: 496 PRRGR-PPVRRGRS-SSFSDSPSP 431
PR R PP+RRGRS SS+S SPSP
Sbjct: 271 PRTRRGPPLRRGRSDSSYSRSPSP 294
Score = 73.9 bits (180), Expect = 2e-12
Identities = 54/125 (43%), Positives = 59/125 (47%), Gaps = 8/125 (6%)
Frame = -2
Query: 658 RGSPGRRRSPPPPPRRRSPPPRRARSPPRR-SPIGRRRSRSPIRRPARSNSRSFSPRRGR 482
R P RR P PPR+R+PP RR SP R+ P RRR SP P R + + RRG
Sbjct: 126 RREPSPRRKPASPPRKRTPPNRRIESPRRQPDPSPRRRPDSP---PIRRRADASPVRRGD 182
Query: 481 PPVRRGRSSSF---SDSPSPRKVSRRSRSRSPRRPLRGG----RASSNSSSSSSPPPPPP 323
P RR S S SP PR R RSP R LRG R S SPPPPP
Sbjct: 183 TPPRRRPGSPVRRRSPSPPPR------RRRSPMRRLRGSPSPRRRSPGPIRRRSPPPPPR 236
Query: 322 PARKP 308
R P
Sbjct: 237 RPRSP 241
Score = 73.2 bits (178), Expect = 3e-12
Identities = 57/137 (41%), Positives = 63/137 (45%), Gaps = 19/137 (13%)
Frame = -2
Query: 673 SPRRLRGSPGRRRSPP-----PPPRRRSPPPRRAR-SPPRRSPIGRRRSRSPIRR----- 527
SPRR SP R+R+PP P R+ P PRR SP PI RR SP+RR
Sbjct: 130 SPRRKPASPPRKRTPPNRRIESPRRQPDPSPRRRPDSP----PIRRRADASPVRRGDTPP 185
Query: 526 ------PARSNSRSFSPRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLR--GGR 371
P R S S PRR R P+RR R S SP + RRS PRRP G R
Sbjct: 186 RRRPGSPVRRRSPSPPPRRRRSPMRRLRGSPSPRRRSPGPIRRRSPPPPPRRPRSPPGRR 245
Query: 370 ASSNSSSSSSPPPPPPP 320
S SPPP PP
Sbjct: 246 LPPPRRHSRSPPPRRPP 262
Score = 39.7 bits (91), Expect = 0.038
Identities = 36/128 (28%), Positives = 49/128 (38%)
Frame = -2
Query: 676 VSPRRLRGSPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGRRRSRSPIRRPARSNSRSFS 497
++PR+ SP ++PPPPP+R P + +P + +RR SP R+PA S
Sbjct: 88 LAPRQRASSP--MKAPPPPPKRDVPHNEKG-APSAEKDVQQRREPSPRRKPA-------S 137
Query: 496 PRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPPPPPPA 317
P R R P R R SPRR P P P
Sbjct: 138 PPRKRTPPNR-------------------RIESPRRQ-----------------PDPSPR 161
Query: 316 RKP*RIPV 293
R+P P+
Sbjct: 162 RRPDSPPI 169
>ref|NP_492875.2| pre-mRNA splicing SR protein related RSR-1 (68.2 kD) (rsr-1)
[Caenorhabditis elegans] gi|19571645|emb|CAB04214.3| C.
elegans RSR-1 protein (corresponding sequence F28D9.1)
[Caenorhabditis elegans]
Length = 601
Score = 82.8 bits (203), Expect = 4e-15
Identities = 57/128 (44%), Positives = 70/128 (54%), Gaps = 6/128 (4%)
Frame = -2
Query: 673 SPRRLRGSPGRRRSPPPPPRR-----RSPPPRRARSP-PRRSPIGRRRSRSPIRRPARSN 512
+P R R SP +SPPP P+R +SPP RR RSP +SP RR RSP S
Sbjct: 343 APARRRRSPSASKSPPPAPKRAKSRSKSPPARRRRSPSASKSPPAPRRRRSP------SK 396
Query: 511 SRSFSPRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPP 332
SRS +P+R PP RR RS S S SP P +SRS+SP P R R S + S S +P
Sbjct: 397 SRSPAPKREIPPARRRRSPSASKSP-PAPKRAKSRSKSPPAPRR--RRSPSQSKSPAPRR 453
Query: 331 PPPPARKP 308
P++ P
Sbjct: 454 RRSPSKSP 461
Score = 82.4 bits (202), Expect = 5e-15
Identities = 60/128 (46%), Positives = 70/128 (53%), Gaps = 5/128 (3%)
Frame = -2
Query: 670 PRRLRGSPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGRRR-----SRSPIRRPARSNSR 506
P R R SP +SPP P RRRSP R+ +P R P RRR S+SP P R+ SR
Sbjct: 372 PARRRRSPSASKSPPAPRRRRSPSKSRSPAPKREIPPARRRRSPSASKSP-PAPKRAKSR 430
Query: 505 SFSPRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPPPP 326
S SP P RR RS S S SP+PR+ RRS S+SP+ P R R S + S S SP
Sbjct: 431 SKSP----PAPRRRRSPSQSKSPAPRR--RRSPSKSPQAPRR--RRSPSGSKSRSPRRRR 482
Query: 325 PPARKP*R 302
PA P R
Sbjct: 483 SPAAAPRR 490
Score = 78.2 bits (191), Expect = 1e-13
Identities = 64/156 (41%), Positives = 78/156 (49%), Gaps = 29/156 (18%)
Frame = -2
Query: 676 VSPRRLRGSPGRRRSP-----PPPPRRR-------SPPPRRARSPPRRSPIGRRR----- 548
V+ R GSP RRRSP PPP RRR SP P+RA+S + P RR
Sbjct: 293 VAARAKSGSPRRRRSPSASKSPPPARRRRSPSQSKSPAPKRAKSRSKSPPAPARRRRSPS 352
Query: 547 -SRSPIRRPARSNSRSFSPRRGRPPVRRGRSSSFSDS-PSPRKVSRRSRSRS--PRRPLR 380
S+SP P R+ SRS S PP RR RS S S S P+PR+ S+SRS P+R +
Sbjct: 353 ASKSPPPAPKRAKSRSKS-----PPARRRRSPSASKSPPAPRRRRSPSKSRSPAPKREIP 407
Query: 379 GGRASSNSSSSSSPPPP--------PPPARKP*RIP 296
R + S+S SPP P PPA + R P
Sbjct: 408 PARRRRSPSASKSPPAPKRAKSRSKSPPAPRRRRSP 443
Score = 72.8 bits (177), Expect = 4e-12
Identities = 52/126 (41%), Positives = 63/126 (49%), Gaps = 5/126 (3%)
Frame = -2
Query: 673 SPRRLRGSPGRRRSPPPPPRRRSP-----PPRRARSPPRRSPIGRRRSRSPIRRPARSNS 509
SP + + R +SPP P RRRSP P R R P +SP RR RSP S S
Sbjct: 420 SPPAPKRAKSRSKSPPAPRRRRSPSQSKSPAPRRRRSPSKSPQAPRRRRSP------SGS 473
Query: 508 RSFSPRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPPP 329
+S SPRR R P + +PR+ R RSPRR R+ S+SS S SPPPP
Sbjct: 474 KSRSPRRRRSP-----------AAAPRRRQSPQRRRSPRR----RRSPSSSSRSRSPPPP 518
Query: 328 PPPARK 311
P R+
Sbjct: 519 PRRPRQ 524
Score = 64.3 bits (155), Expect = 1e-09
Identities = 60/163 (36%), Positives = 76/163 (45%), Gaps = 42/163 (25%)
Frame = -2
Query: 673 SPRRLRGSPGRRRSPPPPPRRRSPP----PRRARSPPRRSPI-----------------G 557
SPRR GSP R PP RR SPP RR R RR P+
Sbjct: 221 SPRR--GSPRRDTRRSPPRRRGSPPARGGDRRDRREDRR-PVRDEKEREEESRRREIERK 277
Query: 556 RRRSRSPIRRPA----RSNSRSFSPRRGR--------PPVRRGRSSSFSDSPSPRKVSRR 413
+R + +R+ A + ++S SPRR R PP RR RS S S SP+P++ +
Sbjct: 278 KRHEEAELRQKAIASVAARAKSGSPRRRRSPSASKSPPPARRRRSPSQSKSPAPKRA--K 335
Query: 412 SRSRSPRRPLRGGRASSNSSSSSSPPPPP---------PPARK 311
SRS+SP P R R + S+S SPPP P PPAR+
Sbjct: 336 SRSKSPPAPARRRR---SPSASKSPPPAPKRAKSRSKSPPARR 375
Score = 51.6 bits (122), Expect = 1e-05
Identities = 48/140 (34%), Positives = 56/140 (39%), Gaps = 17/140 (12%)
Frame = -2
Query: 664 RLRGSPGRRRSPPP---PPRRRSP-------PPRRARSPPRRSPIGRRRSRSPIRRPARS 515
R GS G RR PP PRR SP PPRR SPP R G RR R RRP R
Sbjct: 204 RRGGSAGNRRRSPPRRGSPRRGSPRRDTRRSPPRRRGSPPARG--GDRRDRREDRRPVRD 261
Query: 514 NSRSFSPRRGRPPVRRGRSSSFS-DSPSPRKVSRRSRSRSPRRPLRGGRASS------NS 356
R R R+ R + V+ R++S SPRR + S
Sbjct: 262 EKEREEESRRREIERKKRHEEAELRQKAIASVAARAKSGSPRRRRSPSASKSPPPARRRR 321
Query: 355 SSSSSPPPPPPPARKP*RIP 296
S S S P P A+ + P
Sbjct: 322 SPSQSKSPAPKRAKSRSKSP 341
Score = 51.2 bits (121), Expect = 1e-05
Identities = 42/105 (40%), Positives = 50/105 (47%), Gaps = 10/105 (9%)
Frame = -2
Query: 673 SPRRLRG-------SPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGRRRSRS---PIRRP 524
+PRR R SP RRRSP PRRR P RR RRSP RSRS P RRP
Sbjct: 463 APRRRRSPSGSKSRSPRRRRSPAAAPRRRQSPQRRRSPRRRRSPSSSSRSRSPPPPPRRP 522
Query: 523 ARSNSRSFSPRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRR 389
R +S +P + P + R + +DS VS S + RR
Sbjct: 523 -RQDSEQQAPVAVKSPEMKKRRPNDTDS----DVSVDSEAEEERR 562
Score = 44.7 bits (104), Expect = 0.001
Identities = 47/151 (31%), Positives = 56/151 (36%), Gaps = 29/151 (19%)
Frame = -2
Query: 676 VSPRRLRGSPGRRRSPP--------PPPRRRSPPPRRARSPPRRSPIGRRRSRSPIRRPA 521
+SPR+ +P RR + P P RR R RSPPRR G R SP R
Sbjct: 179 LSPRK---TPPRRNASPGGDGGGGSPAARRGGSAGNRRRSPPRR---GSPRRGSPRRDTR 232
Query: 520 RSNSRSFSPRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLR------------- 380
RS R RRG PP R G + P + + S RR +
Sbjct: 233 RSPPR----RRGSPPARGGDRRDRREDRRPVRDEKEREEESRRREIERKKRHEEAELRQK 288
Query: 379 -----GGRASSNS---SSSSSPPPPPPPARK 311
RA S S S S PPPAR+
Sbjct: 289 AIASVAARAKSGSPRRRRSPSASKSPPPARR 319
Score = 41.2 bits (95), Expect = 0.013
Identities = 31/108 (28%), Positives = 46/108 (41%), Gaps = 2/108 (1%)
Frame = -2
Query: 673 SPRRLRG--SPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGRRRSRSPIRRPARSNSRSF 500
SPRR R S R RSPPPPPRR PR + +++P+ + RRP ++S
Sbjct: 499 SPRRRRSPSSSSRSRSPPPPPRR----PR--QDSEQQAPVAVKSPEMKKRRPNDTDSDVS 552
Query: 499 SPRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNS 356
RR ++ + K ++S+ RR + S S
Sbjct: 553 VDSEAEEERRRKKAKKEKKAAKKHKKEKKSKKHRKRRSPSASESGSES 600
>ref|NP_058079.1| Ser/Arg-related nuclear matrix protein; plenty-of-prolines-101;
serine/arginine repetitive matrix protein 1 [Mus
musculus] gi|3153821|gb|AAC17422.1|
plenty-of-prolines-101; POP101; SH3-philo-protein [Mus
musculus]
Length = 897
Score = 73.6 bits (179), Expect = 2e-12
Identities = 54/135 (40%), Positives = 66/135 (48%), Gaps = 15/135 (11%)
Frame = -2
Query: 670 PRRLRGS--PGRRRSPPPPPRRRSPPPRRARSPPRR----SPIGRRRSRSPIRRPARSNS 509
PRR R P RRR+P PPPRRRSP PRR P +R SP +RR+ SP P R S
Sbjct: 586 PRRRRSPTPPPRRRTPSPPPRRRSPSPRRYSPPIQRRYSPSPPPKRRTASPPPPPKRRAS 645
Query: 508 RSFSPRR--GRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRA-SSNSSSSSSP 338
S P+R P + RS + + SP S+ + SP R R R+ N S SP
Sbjct: 646 PSPPPKRRVSHSPPPKQRSPTVTKRRSPSLSSKHRKGSSPGRSTREARSPQPNKRHSPSP 705
Query: 337 PP------PPPPARK 311
P PPP R+
Sbjct: 706 RPRAPQTSSPPPVRR 720
Score = 66.2 bits (160), Expect = 4e-10
Identities = 52/128 (40%), Positives = 62/128 (47%), Gaps = 7/128 (5%)
Frame = -2
Query: 670 PRRLRGSPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGRRRSRSPIRRPARSNSRSFSPR 491
P R R SP PPPPP PPPRR RSP +P RRR+ SP R RS SPR
Sbjct: 566 PARRRRSPSPAPPPPPPP----PPPRRRRSP---TPPPRRRTPSPPPR-----RRSPSPR 613
Query: 490 RGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPP------- 332
R PP++R +S SP P++ R+ SP P + +S SPPP
Sbjct: 614 RYSPPIQR----RYSPSPPPKR-----RTASPPPPPK-------RRASPSPPPKRRVSHS 657
Query: 331 PPPPARKP 308
PPP R P
Sbjct: 658 PPPKQRSP 665
Score = 65.5 bits (158), Expect = 7e-10
Identities = 57/145 (39%), Positives = 68/145 (46%), Gaps = 19/145 (13%)
Frame = -2
Query: 673 SPRRLRGSPGRRRSPPPPPRRRS----PPPRRARSPPRRSPIGRRR--------SRSPIR 530
SPRR RR SP PPP+RR+ PPP+R SP SP +RR RSP
Sbjct: 611 SPRRYSPPIQRRYSPSPPPKRRTASPPPPPKRRASP---SPPPKRRVSHSPPPKQRSPTV 667
Query: 529 RPARSNSRSFSPRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRR-------PLRGGR 371
RS S S R+G P GRS+ + SP P K R S S PR P+R G
Sbjct: 668 TKRRSPSLSSKHRKGSSP---GRSTREARSPQPNK--RHSPSPRPRAPQTSSPPPVRRG- 721
Query: 370 ASSNSSSSSSPPPPPPPARKP*RIP 296
AS++ SP P P R+ R P
Sbjct: 722 ASASPQGRQSPSPSTRPIRRVSRTP 746
Score = 62.4 bits (150), Expect = 6e-09
Identities = 53/135 (39%), Positives = 59/135 (43%), Gaps = 29/135 (21%)
Frame = -2
Query: 631 PPPPPRRRSPPPR----------RARSPPRRSPIGRRRSRSPI----RRPARSNSRSFSP 494
P P P + P P R RS R R RSRSP RR RS SRS+SP
Sbjct: 250 PEPVPEPKEPSPEKNSKKEKEKTRPRSRSRSKSRSRTRSRSPSHTRPRRRHRSRSRSYSP 309
Query: 493 RRGRPPVRRGRSSSFSDSPSPRKVS---------RRSRSRSPRRPLR------GGRASSN 359
RR P RR PSPR+ + R RSRSP R R G +SS+
Sbjct: 310 RRRPSPRRR---------PSPRRRTPPRRMPPPPRHRRSRSPGRRRRRSSASLSGSSSSS 360
Query: 358 SSSSSSPPPPPPPAR 314
SSS S PP PP R
Sbjct: 361 SSSRSRSPPKKPPKR 375
Score = 60.1 bits (144), Expect = 3e-08
Identities = 62/165 (37%), Positives = 78/165 (46%), Gaps = 49/165 (29%)
Frame = -2
Query: 667 RRLRGSPGRRR---SPPPPP-RRR--SPPPRR--ARSPP--RRSP-IGRRRSRSPIRR-- 527
RR SP +R SPPPPP RR SPPP+R + SPP +RSP + +RRS S +
Sbjct: 621 RRYSPSPPPKRRTASPPPPPKRRASPSPPPKRRVSHSPPPKQRSPTVTKRRSPSLSSKHR 680
Query: 526 ----PARSNSRSFSPRRGR----------------PPVRRGRSSS----FSDSPSPRKVS 419
P RS + SP+ + PPVRRG S+S S SPS R +
Sbjct: 681 KGSSPGRSTREARSPQPNKRHSPSPRPRAPQTSSPPPVRRGASASPQGRQSPSPSTRPIR 740
Query: 418 RRSRSRSPRR--------PLRGGRASSNSSSSSSPPP----PPPP 320
R SR+ P++ P R SS+ S S SP P PP P
Sbjct: 741 RVSRTPEPKKIKKAASPSPQSVRRVSSSRSVSGSPEPAAKKPPAP 785
Score = 58.9 bits (141), Expect = 6e-08
Identities = 52/119 (43%), Positives = 55/119 (45%), Gaps = 6/119 (5%)
Frame = -2
Query: 652 SPGRRRSP--PPPPRRRSPPPRRARSPPR----RSPIGRRRSRSPIRRPARSNSRSFSPR 491
SP RR SP P PRRR+ PPRR PPR RSP GRRR RS S+S S S
Sbjct: 308 SPRRRPSPRRRPSPRRRT-PPRRMPPPPRHRRSRSP-GRRRRRSSASLSGSSSSSSSSRS 365
Query: 490 RGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPPPPPPAR 314
R P R+SS PRK R S S SP R SSP PPP R
Sbjct: 366 RSPPKKPPKRTSS-----PPRKTRRLSPSASPPR---------RRHRPSSPATPPPKTR 410
Score = 58.5 bits (140), Expect = 8e-08
Identities = 55/139 (39%), Positives = 62/139 (44%), Gaps = 14/139 (10%)
Frame = -2
Query: 673 SPRRLRGSPGRRRSPPPPPRRRSPPPRRARSP----PRRSPIGRRRSRSPIRRPARSNSR 506
SP + + P R +S R+RSP PRR R RS SP RRP S R
Sbjct: 260 SPEKNSKKEKEKTRPRSRSRSKSRSRTRSRSPSHTRPRRRHRSRSRSYSPRRRP--SPRR 317
Query: 505 SFSPRRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRR-----PLRGGRASSNSSSSSS 341
SPRR PP R P PR RRSRS RR L G +SS+SS S S
Sbjct: 318 RPSPRRRTPPRRM--------PPPPR--HRRSRSPGRRRRRSSASLSGSSSSSSSSRSRS 367
Query: 340 PPPPPP-----PARKP*RI 299
PP PP P RK R+
Sbjct: 368 PPKKPPKRTSSPPRKTRRL 386
Score = 57.4 bits (137), Expect = 2e-07
Identities = 56/152 (36%), Positives = 64/152 (41%), Gaps = 37/152 (24%)
Frame = -2
Query: 652 SPGRRRSPPPPPRRRSPPPRRARSPPR-RSPIGRRRSRSPIRRPARSNSRSFSPRRGRPP 476
SP RR+S P PRRRS P RR R RSP R +SRSP P + SP P
Sbjct: 171 SPRRRKSRSPSPRRRSSPVRRERKRSHSRSPRHRTKSRSPSPAPEKKEK---SPELPEPS 227
Query: 475 VRRGRSS-----SFSD--------------SPSPRKVSR--------RSRSRSPRRPLRG 377
VR SS S SD PSP K S+ RSRSRS R
Sbjct: 228 VRMKDSSVQEATSTSDILKAPKPEPVPEPKEPSPEKNSKKEKEKTRPRSRSRSKSRSRTR 287
Query: 376 GRASSNS---------SSSSSPPPPPPPARKP 308
R+ S++ S S SP P P R+P
Sbjct: 288 SRSPSHTRPRRRHRSRSRSYSPRRRPSPRRRP 319
Score = 52.0 bits (123), Expect = 7e-06
Identities = 51/130 (39%), Positives = 58/130 (44%), Gaps = 9/130 (6%)
Frame = -2
Query: 670 PRRLRGSPGRRRSPP--PPPRRRSPPPRRARSPPRRSPIGRRRSRSPIRRPARSNSRSFS 497
PRR S R SP P PRRR P PRR R+PPRR P P + RS S
Sbjct: 296 PRRRHRSRSRSYSPRRRPSPRRR-PSPRR-RTPPRRMP------------PPPRHRRSRS 341
Query: 496 PRRGRPPVRRGRSSSFSDSPSPRKVSRRSRS---RSPRRPLRGGRASSNSSSSSSPP--- 335
P R RR RSS+ S S RSRS + P+R R + S S+SPP
Sbjct: 342 PGR-----RRRRSSASLSGSSSSSSSSRSRSPPKKPPKRTSSPPRKTRRLSPSASPPRRR 396
Query: 334 -PPPPPARKP 308
P PA P
Sbjct: 397 HRPSSPATPP 406
Score = 49.7 bits (117), Expect = 4e-05
Identities = 48/133 (36%), Positives = 55/133 (41%), Gaps = 14/133 (10%)
Frame = -2
Query: 667 RRLRGSPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGR--RRSRSPIRRPARSNSRSFSP 494
RR R SP R SP P R++ PR +SP+ + RR RSP SP
Sbjct: 517 RRRRLSPSRSASPSPRKRQKETSPRMQMGKRWQSPVTKSSRRRRSP------------SP 564
Query: 493 RRGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSP------RRPLRGGRASSNSSSSSSPP- 335
PP RR RS S + P P R R RSP R P R S S SPP
Sbjct: 565 ----PPARRRRSPSPAPPPPPPPPPPRRR-RSPTPPPRRRTPSPPPRRRSPSPRRYSPPI 619
Query: 334 -----PPPPPARK 311
P PPP R+
Sbjct: 620 QRRYSPSPPPKRR 632
Score = 48.1 bits (113), Expect = 1e-04
Identities = 41/109 (37%), Positives = 47/109 (42%), Gaps = 14/109 (12%)
Frame = -2
Query: 595 RRARSPPRRSPIGRRRSRSPIRRPARSNSRSFSPRRGRPPVRRGRSSSFSDSPSPRKVSR 416
+R + S R RSRSP RR SRS SPRR PVRR R S S SP R SR
Sbjct: 153 KRDKEEKESSREKRERSRSPRRR----KSRSPSPRRRSSPVRRERKRSHSRSPRHRTKSR 208
Query: 415 ------RSRSRSPRRPLRGGR--------ASSNSSSSSSPPPPPPPARK 311
+ +SP P R A+S S +P P P P K
Sbjct: 209 SPSPAPEKKEKSPELPEPSVRMKDSSVQEATSTSDILKAPKPEPVPEPK 257
Score = 47.0 bits (110), Expect = 2e-04
Identities = 41/130 (31%), Positives = 51/130 (38%), Gaps = 8/130 (6%)
Frame = -2
Query: 676 VSPRRLRGSPGRRRSPPPPPRRRSPPPRRARSPPRRSPIGRRRSRSPIRRPA---RSNSR 506
V RR ++ S S R RS + +GRRR SP R + R +
Sbjct: 477 VQQRRQYRRQNQQSSSDSGSSSTSEDERPKRSHVKNGEVGRRRRLSPSRSASPSPRKRQK 536
Query: 505 SFSPR-----RGRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSS 341
SPR R + PV + S SP P +RR RS SP P
Sbjct: 537 ETSPRMQMGKRWQSPVTKSSRRRRSPSPPP---ARRRRSPSPAPP--------------P 579
Query: 340 PPPPPPPARK 311
PPPPPPP R+
Sbjct: 580 PPPPPPPRRR 589
Score = 43.9 bits (102), Expect = 0.002
Identities = 39/118 (33%), Positives = 53/118 (44%), Gaps = 6/118 (5%)
Frame = -2
Query: 649 PGRRRSPPPPPRR---RSPPPRRARSPPRRSPIGRRR---SRSPIRRPARSNSRSFSPRR 488
P +R SP P PR SPPP R SP GR+ S PIRR +R+
Sbjct: 697 PNKRHSPSPRPRAPQTSSPPP--VRRGASASPQGRQSPSPSTRPIRRVSRT--------- 745
Query: 487 GRPPVRRGRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPPPPPPAR 314
P ++ + ++ SPSP+ V R S SRS + S ++ PP PP P +
Sbjct: 746 --PEPKKIKKAA---SPSPQSVRRVSSSRSV--------SGSPEPAAKKPPAPPSPVQ 790
Score = 43.5 bits (101), Expect = 0.003
Identities = 46/147 (31%), Positives = 58/147 (39%), Gaps = 34/147 (23%)
Frame = -2
Query: 652 SPGRRRSPP--PPPRRRSPPPRRARSPPRRSPIGRR-RSRSPIRRPARSN-----SRSFS 497
S R RSPP PP R SPP + R P SP RR R SP P ++ +S
Sbjct: 361 SSSRSRSPPKKPPKRTSSPPRKTRRLSPSASPPRRRHRPSSPATPPPKTRHSPTPQQSNR 420
Query: 496 PRRGRPPVRRGRSS-------------SFSDSPSPRKV-------------SRRSRSRSP 395
R+ R V GR+S S S +P PRKV + +
Sbjct: 421 TRKSRVSVSPGRTSGKVTKHKGTEKRESPSPAPKPRKVELSESEEDKGSKMAAADSVQQR 480
Query: 394 RRPLRGGRASSNSSSSSSPPPPPPPAR 314
R+ R + SS+ S SSS P R
Sbjct: 481 RQYRRQNQQSSSDSGSSSTSEDERPKR 507
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 608,802,912
Number of Sequences: 1393205
Number of extensions: 15520371
Number of successful extensions: 286168
Number of sequences better than 10.0: 6327
Number of HSP's better than 10.0 without gapping: 101147
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 204873
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29987172312
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)