Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002179A_C01 KMC002179A_c01
(502 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
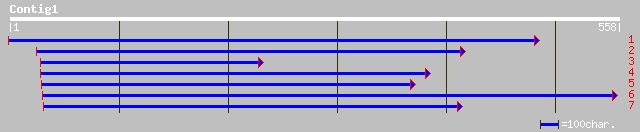
emb|CAC84706.1| aux/IAA protein [Populus tremula x Populus tremu... 83 2e-15
dbj|BAA85821.1| CS-IAA2 [Cucumis sativus] 83 2e-15
pir||T05726 GH1 protein - soybean (fragment) gi|2388689|gb|AAB70... 82 3e-15
gb|AAM12952.1| auxin-regulated protein [Zinnia elegans] 81 8e-15
gb|AAM29182.1| Aux/IAA protein [Solanum tuberosum] 80 1e-14
>emb|CAC84706.1| aux/IAA protein [Populus tremula x Populus tremuloides]
Length = 365
Score = 83.2 bits (204), Expect = 2e-15
Identities = 42/71 (59%), Positives = 55/71 (77%), Gaps = 3/71 (4%)
Frame = -2
Query: 501 EIMNETKLKDLIHGSENVLTYEDKDGELEALWVM---FPGRCLLIACRRLRIMKSSDALG 331
E+++E+KLKDL+HGSE VLTYEDKDG+ W++ P + C+RLRIMKSSDA+G
Sbjct: 297 EMLSESKLKDLLHGSEYVLTYEDKDGD----WMLVGDVPWEMFIETCKRLRIMKSSDAIG 352
Query: 330 LAPRAVEKSKS 298
LAPRA+EK K+
Sbjct: 353 LAPRAMEKCKN 363
>dbj|BAA85821.1| CS-IAA2 [Cucumis sativus]
Length = 355
Score = 83.2 bits (204), Expect = 2e-15
Identities = 43/71 (60%), Positives = 55/71 (76%), Gaps = 3/71 (4%)
Frame = -2
Query: 501 EIMNETKLKDLIHGSENVLTYEDKDGELEALWVM---FPGRCLLIACRRLRIMKSSDALG 331
E M+E+KLKDL+HGSE VLTYEDKDG+ W++ P + +C+RLRIMKSSDA+G
Sbjct: 287 EKMSESKLKDLLHGSEYVLTYEDKDGD----WMLVGDVPWEMFIDSCKRLRIMKSSDAIG 342
Query: 330 LAPRAVEKSKS 298
LAPRAVEK ++
Sbjct: 343 LAPRAVEKCRN 353
>pir||T05726 GH1 protein - soybean (fragment) gi|2388689|gb|AAB70005.1| GH1
protein [Glycine max]
Length = 339
Score = 82.4 bits (202), Expect = 3e-15
Identities = 41/71 (57%), Positives = 55/71 (76%), Gaps = 3/71 (4%)
Frame = -2
Query: 501 EIMNETKLKDLIHGSENVLTYEDKDGELEALWVMF---PGRCLLIACRRLRIMKSSDALG 331
E+++E+KL+DL+HGSE VLTYEDKDG+ W++ P + C+RL+IMK SDA+G
Sbjct: 271 EMLSESKLRDLLHGSEYVLTYEDKDGD----WMLVGDVPWEMFIDTCKRLKIMKGSDAIG 326
Query: 330 LAPRAVEKSKS 298
LAPRA+EKSKS
Sbjct: 327 LAPRAMEKSKS 337
>gb|AAM12952.1| auxin-regulated protein [Zinnia elegans]
Length = 351
Score = 80.9 bits (198), Expect = 8e-15
Identities = 42/72 (58%), Positives = 55/72 (76%), Gaps = 4/72 (5%)
Frame = -2
Query: 501 EIMNETKLKDLIHGSENVLTYEDKDGEL----EALWVMFPGRCLLIACRRLRIMKSSDAL 334
E ++E+KL+DL+HGSE VLTYEDKDG+ + W MF G +C+RL+IMK SDA+
Sbjct: 283 ESLSESKLRDLLHGSEYVLTYEDKDGDWMLVGDVPWDMFIG-----SCKRLKIMKGSDAI 337
Query: 333 GLAPRAVEKSKS 298
GLAPRA+EKSK+
Sbjct: 338 GLAPRAMEKSKN 349
>gb|AAM29182.1| Aux/IAA protein [Solanum tuberosum]
Length = 349
Score = 80.5 bits (197), Expect = 1e-14
Identities = 40/71 (56%), Positives = 54/71 (75%), Gaps = 3/71 (4%)
Frame = -2
Query: 501 EIMNETKLKDLIHGSENVLTYEDKDGELEALWVM---FPGRCLLIACRRLRIMKSSDALG 331
++++E+KLKDL+HGSE VLTYEDKDG+ W++ P + C+RLRIMK SDA+G
Sbjct: 281 DMLSESKLKDLLHGSEYVLTYEDKDGD----WMLVGDVPWEMFIDTCKRLRIMKGSDAIG 336
Query: 330 LAPRAVEKSKS 298
LAPRA+EK +S
Sbjct: 337 LAPRAMEKCRS 347
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 427,707,851
Number of Sequences: 1393205
Number of extensions: 8616629
Number of successful extensions: 21201
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 19984
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20909
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 15072921604
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)