Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
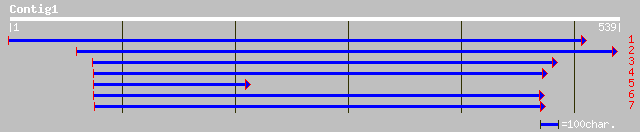
Query= KMC002162A_C01 KMC002162A_c01
(450 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_195822.1| putative protein; protein id: At5g02020.1, supp... 60 6e-09
gb|AAO37201.1| hypothetical protein [Arabidopsis thaliana] 44 4e-04
ref|NP_671943.1| hypothetical protein; protein id: At2g39855.1 [... 42 0.001
ref|NP_200716.1| putative protein; protein id: At5g59080.1, supp... 38 0.024
ref|NP_680147.1| hypothetical protein; protein id: At5g05965.1 [... 35 0.27
>ref|NP_195822.1| putative protein; protein id: At5g02020.1, supported by cDNA:
gi_16648747, supported by cDNA: gi_20334905 [Arabidopsis
thaliana] gi|11358413|pir||T48223 hypothetical protein
T7H20.70 - Arabidopsis thaliana
gi|7340676|emb|CAB82975.1| putative protein [Arabidopsis
thaliana] gi|16648748|gb|AAL25566.1| AT5g02020/T7H20_70
[Arabidopsis thaliana] gi|20334906|gb|AAM16209.1|
AT5g02020/T7H20_70 [Arabidopsis thaliana]
Length = 149
Score = 60.1 bits (144), Expect = 6e-09
Identities = 26/33 (78%), Positives = 28/33 (84%)
Frame = -3
Query: 445 RDSRLNSLYKNDGGEDDSGSASRGNWWQGSLYY 347
++S NS K DGGEDDSGSASRGNWWQGSLYY
Sbjct: 117 QNSTSNSTNKKDGGEDDSGSASRGNWWQGSLYY 149
>gb|AAO37201.1| hypothetical protein [Arabidopsis thaliana]
Length = 142
Score = 43.9 bits (102), Expect = 4e-04
Identities = 18/25 (72%), Positives = 19/25 (76%)
Frame = -3
Query: 421 YKNDGGEDDSGSASRGNWWQGSLYY 347
YK DG E DS SASRGNWW+GS Y
Sbjct: 118 YKKDGEEGDSESASRGNWWEGSFNY 142
>ref|NP_671943.1| hypothetical protein; protein id: At2g39855.1 [Arabidopsis
thaliana]
Length = 155
Score = 42.4 bits (98), Expect = 0.001
Identities = 17/23 (73%), Positives = 19/23 (81%)
Frame = -3
Query: 421 YKNDGGEDDSGSASRGNWWQGSL 353
YK DG E DS SASRGNWW+G+L
Sbjct: 131 YKKDGEEGDSESASRGNWWEGTL 153
>ref|NP_200716.1| putative protein; protein id: At5g59080.1, supported by cDNA:
gi_17380959, supported by cDNA: gi_20465392 [Arabidopsis
thaliana] gi|10177635|dbj|BAB10783.1|
emb|CAB82975.1~gene_id:K18B18.8~similar to unknown
protein [Arabidopsis thaliana]
gi|17380960|gb|AAL36292.1| unknown protein [Arabidopsis
thaliana] gi|20465393|gb|AAM20121.1| unknown protein
[Arabidopsis thaliana]
Length = 135
Score = 38.1 bits (87), Expect = 0.024
Identities = 19/32 (59%), Positives = 20/32 (62%), Gaps = 8/32 (25%)
Frame = -3
Query: 418 KND---GGEDDSGS-----ASRGNWWQGSLYY 347
KND GEDD+ SRGNWWQGSLYY
Sbjct: 104 KNDRRRSGEDDANGQNPQDVSRGNWWQGSLYY 135
>ref|NP_680147.1| hypothetical protein; protein id: At5g05965.1 [Arabidopsis
thaliana]
Length = 131
Score = 34.7 bits (78), Expect = 0.27
Identities = 13/18 (72%), Positives = 15/18 (83%)
Frame = -3
Query: 400 DDSGSASRGNWWQGSLYY 347
D + S SRGNWW+GSLYY
Sbjct: 114 DITTSTSRGNWWKGSLYY 131
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 379,399,033
Number of Sequences: 1393205
Number of extensions: 7582306
Number of successful extensions: 15994
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 15716
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 15993
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 6588926928
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)