Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002155A_C01 KMC002155A_c01
(967 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
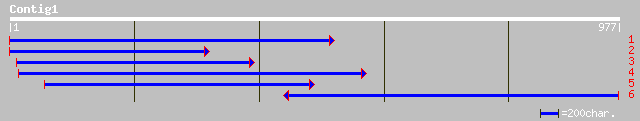
ref|NP_190351.1| putative protein; protein id: At3g47670.1, supp... 137 3e-31
sp|Q43111|PME3_PHAVU Pectinesterase 3 precursor (Pectin methyles... 132 6e-30
gb|AAM65650.1| pectinesterase, putative [Arabidopsis thaliana] 127 2e-28
ref|NP_175787.1| pectinesterase family; protein id: At1g53840.1,... 127 2e-28
ref|NP_188047.1| putative pectin methylesterase; protein id: At3... 110 2e-23
>ref|NP_190351.1| putative protein; protein id: At3g47670.1, supported by cDNA:
gi_14517535 [Arabidopsis thaliana]
gi|11357584|pir||T45727 hypothetical protein F1P2.220 -
Arabidopsis thaliana gi|6522550|emb|CAB61993.1| putative
protein [Arabidopsis thaliana]
gi|14517536|gb|AAK62658.1| AT3g47670/F1P2_220
[Arabidopsis thaliana] gi|23308197|gb|AAN18068.1|
At3g47670/F1P2_220 [Arabidopsis thaliana]
Length = 240
Score = 137 bits (344), Expect = 3e-31
Identities = 80/229 (34%), Positives = 132/229 (56%), Gaps = 6/229 (2%)
Frame = -3
Query: 965 GYGKVEEEEHNHHHHHNLEDQQQRNQHHHRKASTAIISILAILFFTLALAFTLATLLHHT 786
GY KV ++E+ HHHH+ + RK A S++++L +A T H
Sbjct: 9 GYDKVSDQENQIHHHHH--SPLPKPSSFSRKTLIATGSVVSLLLILSVVALTAGAFTH-- 64
Query: 785 TTTTKPPHKQQQPLNSADSIRTVCNVTRFPDSCIAAVSSS---NDHPTDPQSILTLSLRA 615
PPH P++SA S++++C+VTR+P++C ++SSS +D +P+SIL LSLR
Sbjct: 65 ----SPPH--HPPISSA-SLKSLCSVTRYPETCFNSLSSSLNESDSKLNPESILELSLRV 117
Query: 614 SVDELASVASSLGVKASSSNGQGVADCKDQVDDALSRLNDSLSAMAVVP---GGVTLTDA 444
+ E+++++ S V DC DALS+LNDS++ + G LT+
Sbjct: 118 AAKEISNLSISFRSINDMPEDAAVGDCVKLYTDALSQLNDSITEIERKKKKGGNNWLTEE 177
Query: 443 KISDIQTWVSAAVTDQQTCLDGLEEVGSVVVEEVKNMMKKSSEYVSNSL 297
+ D++TW+SAA+TD +TC DG+EE+G++V E+K M+ +++ +S SL
Sbjct: 178 VVGDVKTWISAAMTDGETCSDGIEEMGTIVGNEIKKKMEMANQMMSISL 226
>sp|Q43111|PME3_PHAVU Pectinesterase 3 precursor (Pectin methylesterase 3) (PE 3)
gi|1076515|pir||S53105 pectinesterase precursor - kidney
bean gi|732913|emb|CAA59482.1| pectinesterase [Phaseolus
vulgaris]
Length = 581
Score = 132 bits (333), Expect = 6e-30
Identities = 81/232 (34%), Positives = 128/232 (54%), Gaps = 9/232 (3%)
Frame = -3
Query: 965 GYGKVEEEEHNHHHHHNLEDQQQRNQHHHRKASTAIISILAILFFTLALAFTLATLLHHT 786
GYGKV E E QQ + RK II++ +I+ + +A ++H+
Sbjct: 9 GYGKVNELE------------QQAYEKKTRKR-LIIIAVSSIVLIAVIIAAVAGVVIHNR 55
Query: 785 TTTTKPPHKQ--QQPLNSADSIRTVCNVTRFPDSCIAAVSS-SNDHPTDPQSILTLSLRA 615
+ + P Q L+ A S++ VC+ TR+P SC +++SS + TDP+ + LSLR
Sbjct: 56 NSESSPSSDSVPQTELSPAASLKAVCDTTRYPSSCFSSISSLPESNTTDPELLFKLSLRV 115
Query: 614 SVDELASVASSLGVKASSSNG--QGVADCKDQVDDALSRLNDSLSAMAVVPGGVTLTDAK 441
++DEL+S S L A + + C DAL RLNDS+SA+ V G + + A
Sbjct: 116 AIDELSSFPSKLRANAEQDARLQKAIDVCSSVFGDALDRLNDSISALGTVAGRIA-SSAS 174
Query: 440 ISDIQTWVSAAVTDQQTCLDGLEEVGSV----VVEEVKNMMKKSSEYVSNSL 297
+S+++TW+SAA+TDQ TCLD + E+ S ++E++ M+ S+E+ SNSL
Sbjct: 175 VSNVETWLSAALTDQDTCLDAVGELNSTAARGALQEIETAMRNSTEFASNSL 226
>gb|AAM65650.1| pectinesterase, putative [Arabidopsis thaliana]
Length = 586
Score = 127 bits (320), Expect = 2e-28
Identities = 80/240 (33%), Positives = 134/240 (55%), Gaps = 17/240 (7%)
Frame = -3
Query: 965 GYGKVEEEEHNHHHHHNLEDQQQRNQHHHRKASTAIISILAILFFTLALAFTLATLLH-- 792
GYGKV+E + D + + R ++SI ++ + +A +AT++H
Sbjct: 9 GYGKVDEAQ----------DLALKKKTRKR---LLLLSISVVVLIAVIIAAVVATVVHKN 55
Query: 791 -HTTTTTKPPHKQQQPLNSADSIRTVCNVTRFPDSCIAAVSS-SNDHPTDPQSILTLSLR 618
+ +T + PP L + S++ +C+VTRFP+SCI+++S + + TDP+++ LSL+
Sbjct: 56 KNESTPSPPPE-----LTPSTSLKAICSVTRFPESCISSISKLPSSNTTDPETLFKLSLK 110
Query: 617 ASVDELASVASSLGVKASSSNGQ-----GVADCKDQVDDALSRLNDSLSAMAVVPGGVTL 453
+DEL S+ S L K S + C D ++DAL RLND++SA+ TL
Sbjct: 111 VIIDELDSI-SDLPEKLSKETEDERIKSALRVCGDLIEDALDRLNDTVSAIDDEEKKKTL 169
Query: 452 TDAKISDIQTWVSAAVTDQQTCLDGLEEV--------GSVVVEEVKNMMKKSSEYVSNSL 297
+ +KI D++TW+SA VTD +TC D L+E+ S + + +K+ M +S+E+ SNSL
Sbjct: 170 SSSKIEDLKTWLSATVTDHETCFDSLDELKQNKTEYANSTITQNLKSAMSRSTEFTSNSL 229
>ref|NP_175787.1| pectinesterase family; protein id: At1g53840.1, supported by cDNA:
41374., supported by cDNA: gi_15809859 [Arabidopsis
thaliana] gi|6093736|sp|Q43867|PME1_ARATH Pectinesterase
1 precursor (Pectin methylesterase 1) (PE 1)
gi|2129666|pir||JC4778 pectinesterase (EC 3.1.1.11) PME1
precursor - Arabidopsis thaliana
gi|550306|emb|CAA57275.1| ATPME1 [Arabidopsis thaliana]
gi|903895|gb|AAC50024.1| ATPME1 precursor [Arabidopsis
thaliana] gi|6056393|gb|AAF02857.1|AC009324_6
Pectinesterase 1 [Arabidopsis thaliana]
gi|15809860|gb|AAL06858.1| At1g53840/T18A20_7
[Arabidopsis thaliana]
Length = 586
Score = 127 bits (320), Expect = 2e-28
Identities = 80/240 (33%), Positives = 134/240 (55%), Gaps = 17/240 (7%)
Frame = -3
Query: 965 GYGKVEEEEHNHHHHHNLEDQQQRNQHHHRKASTAIISILAILFFTLALAFTLATLLH-- 792
GYGKV+E + D + + R ++SI ++ + +A +AT++H
Sbjct: 9 GYGKVDEAQ----------DLALKKKTRKR---LLLLSISVVVLIAVIIAAVVATVVHKN 55
Query: 791 -HTTTTTKPPHKQQQPLNSADSIRTVCNVTRFPDSCIAAVSS-SNDHPTDPQSILTLSLR 618
+ +T + PP L + S++ +C+VTRFP+SCI+++S + + TDP+++ LSL+
Sbjct: 56 KNESTPSPPPE-----LTPSTSLKAICSVTRFPESCISSISKLPSSNTTDPETLFKLSLK 110
Query: 617 ASVDELASVASSLGVKASSSNGQ-----GVADCKDQVDDALSRLNDSLSAMAVVPGGVTL 453
+DEL S+ S L K S + C D ++DAL RLND++SA+ TL
Sbjct: 111 VIIDELDSI-SDLPEKLSKETEDERIKSALRVCGDLIEDALDRLNDTVSAIDDEEKKKTL 169
Query: 452 TDAKISDIQTWVSAAVTDQQTCLDGLEEV--------GSVVVEEVKNMMKKSSEYVSNSL 297
+ +KI D++TW+SA VTD +TC D L+E+ S + + +K+ M +S+E+ SNSL
Sbjct: 170 SSSKIEDLKTWLSATVTDHETCFDSLDELKQNKTEYANSTITQNLKSAMSRSTEFTSNSL 229
>ref|NP_188047.1| putative pectin methylesterase; protein id: At3g14300.1
[Arabidopsis thaliana] gi|9279578|dbj|BAB01036.1|
pectinesterase-like protein [Arabidopsis thaliana]
Length = 968
Score = 110 bits (276), Expect = 2e-23
Identities = 75/232 (32%), Positives = 125/232 (53%), Gaps = 9/232 (3%)
Frame = -3
Query: 965 GYGKVEEEEHNHHHHHNLEDQQQRNQHHHRKASTAIISILAILFFTLALAFTLATLLHHT 786
GYGKV+E + +L +++ + ++ + + + I+ T+ +A +H
Sbjct: 10 GYGKVDETQ-------DLALKRKTRKRLYQIGISVAVLVAIIISSTVTIA------IHSR 56
Query: 785 TTTTKPPHKQQQP-LNSADSIRTVCNVTRFPDSCIAAVSSSN-DHPTDPQSILTLSLRAS 612
+ P P L A S++TVC+VT +P SC +++S + TDP+ I LSL+
Sbjct: 57 KGNSPHPTPSSVPELTPAASLKTVCSVTNYPVSCFSSISKLPLSNTTDPEVIFRLSLQVV 116
Query: 611 VDELASVASSLGVKASSSNGQGVAD----CKDQVDDALSRLNDSLSAMAVVPGGVTLTDA 444
+DEL S+ A ++ +G+ C+ +D A+ R+N+++SAM VV G L A
Sbjct: 117 IDELNSIVELPKKLAEETDDEGLKSALSVCEHLLDLAIDRVNETVSAMEVVDGKKILNAA 176
Query: 443 KISDIQTWVSAAVTDQQTCLDGLEEV---GSVVVEEVKNMMKKSSEYVSNSL 297
I D+ TW+SAAVT TCLD L+E+ S + ++K+ M S+E+ SNSL
Sbjct: 177 TIDDLLTWLSAAVTYHGTCLDALDEISHTNSAIPLKLKSGMVNSTEFTSNSL 228
Score = 100 bits (250), Expect = 3e-20
Identities = 67/201 (33%), Positives = 107/201 (52%), Gaps = 17/201 (8%)
Frame = -3
Query: 848 LAILFFTLALAFTLATLLHHTTTTTKPPHKQQ---QPLNSADSIRTVCNVTRFPDSCIAA 678
LAI+ L+ +H PH L A S+R VC+VTR+P SC+++
Sbjct: 228 LAIVAKILSTISDFGIPIHGRRLLNSSPHATPISVPKLTPAASLRNVCSVTRYPASCVSS 287
Query: 677 VSS-SNDHPTDPQSILTLSLRASVDELASVASSLGVKASSSNGQ----GVADCKDQVDDA 513
+S + + TDP+++ LSL+ ++EL S+A A ++ + ++ C D +DA
Sbjct: 288 ISKLPSSNTTDPEALFRLSLQVVINELNSIAGLPKKLAEETDDERLKSSLSVCGDVFNDA 347
Query: 512 LSRLNDSLSAMAVVPGGVT-LTDAKISDIQTWVSAAVTDQQTCLDGLEEV--------GS 360
+ +ND++S M V G L + I +IQTW+SAAVTD TCLD L+E+ S
Sbjct: 348 IDIVNDTISTMEEVGDGKKILKSSTIDEIQTWLSAAVTDHDTCLDALDELSQNKTEYANS 407
Query: 359 VVVEEVKNMMKKSSEYVSNSL 297
+ ++K+ M S ++ SNSL
Sbjct: 408 PISLKLKSAMVNSRKFTSNSL 428
Score = 97.1 bits (240), Expect = 4e-19
Identities = 63/176 (35%), Positives = 100/176 (56%), Gaps = 11/176 (6%)
Frame = -3
Query: 791 HTTTTTKPPHKQQQPLNSADSI-RTVCNVTRFPDSCIAAVSS--SNDHPTDPQSILTLSL 621
H + P ++ P + S+ RTVCNVT +P SCI+++S + TDP+ + LSL
Sbjct: 437 HERHGVQSPRLRKSPHPTPSSVLRTVCNVTNYPASCISSISKLPLSKTTTDPKVLFRLSL 496
Query: 620 RASVDELASVASSLGVKASSSNGQGVAD----CKDQVDDALSRLNDSLSAM-AVVPGG-V 459
+ + DEL S+ A +N +G+ C D D A+ +ND++S++ V+ GG
Sbjct: 497 QVTFDELNSIVGLPKKLAEETNDEGLKSALSVCADVFDLAVDSVNDTISSLDEVISGGKK 556
Query: 458 TLTDAKISDIQTWVSAAVTDQQTCLDGLEE--VGSVVVEEVKNMMKKSSEYVSNSL 297
L + I D+ TW+S+AVTD TC D L+E S + +++K+ M S+E+ SNSL
Sbjct: 557 NLNSSTIGDLITWLSSAVTDIGTCGDTLDEDNYNSPIPQKLKSAMVNSTEFTSNSL 612
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 999,210,977
Number of Sequences: 1393205
Number of extensions: 30217643
Number of successful extensions: 773128
Number of sequences better than 10.0: 16209
Number of HSP's better than 10.0 without gapping: 184926
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 477781
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 54910356336
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)