Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
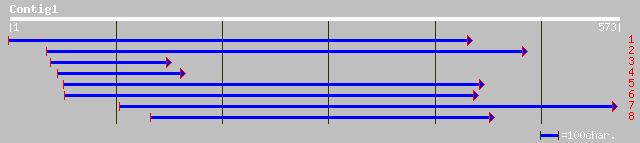
Query= KMC002149A_C01 KMC002149A_c01
(573 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||B84652 hypothetical protein At2g25740 [imported] - Arabidop... 174 7e-43
ref|NP_671881.1| hypothetical protein; protein id: At2g25737.1 [... 174 7e-43
gb|AAL27555.1|AF420410_1 hypothetical protein [Musa acuminata] 151 6e-36
ref|NP_181201.2| unknown protein; protein id: At2g36630.1, suppo... 133 2e-30
gb|AAF16643.1|AC011661_21 T23J18.20 [Arabidopsis thaliana] 82 5e-15
>pir||B84652 hypothetical protein At2g25740 [imported] - Arabidopsis thaliana
gi|3643608|gb|AAC42255.1| hypothetical protein
[Arabidopsis thaliana] gi|20197510|gb|AAM15102.1|
hypothetical protein [Arabidopsis thaliana]
Length = 902
Score = 174 bits (441), Expect = 7e-43
Identities = 87/122 (71%), Positives = 102/122 (83%)
Frame = -3
Query: 571 GGGFVMGPLFLELGIAPQVASATATFGMTFSASISVVQYYLLNRFPVPYALYLTLVAAIA 392
GGGF+MGPLFLELG+ PQV+SATATF MTFS+S+SVV+YYLL RFPVPYALYL VA IA
Sbjct: 208 GGGFIMGPLFLELGVPPQVSSATATFAMTFSSSMSVVEYYLLKRFPVPYALYLVGVATIA 267
Query: 391 AYRGQFIIDKLVNMFQRASLIIFTLAFTIFVSAIVLGGEGISDMIGQIQRSEYMGFEDLC 212
A+ GQ ++ +L+ RASLIIF LA IF+SAI LGG GI +MIG+IQR EYMGFE+LC
Sbjct: 268 AWVGQHVVRRLIAAIGRASLIIFILASMIFISAISLGGVGIVNMIGKIQRHEYMGFENLC 327
Query: 211 KY 206
KY
Sbjct: 328 KY 329
>ref|NP_671881.1| hypothetical protein; protein id: At2g25737.1 [Arabidopsis
thaliana] gi|22531098|gb|AAM97053.1| unknown protein
[Arabidopsis thaliana] gi|25083770|gb|AAN72117.1|
unknown protein [Arabidopsis thaliana]
gi|26450521|dbj|BAC42374.1| unknown protein [Arabidopsis
thaliana] gi|28951043|gb|AAO63445.1| At2g25737
[Arabidopsis thaliana]
Length = 476
Score = 174 bits (441), Expect = 7e-43
Identities = 87/122 (71%), Positives = 102/122 (83%)
Frame = -3
Query: 571 GGGFVMGPLFLELGIAPQVASATATFGMTFSASISVVQYYLLNRFPVPYALYLTLVAAIA 392
GGGF+MGPLFLELG+ PQV+SATATF MTFS+S+SVV+YYLL RFPVPYALYL VA IA
Sbjct: 353 GGGFIMGPLFLELGVPPQVSSATATFAMTFSSSMSVVEYYLLKRFPVPYALYLVGVATIA 412
Query: 391 AYRGQFIIDKLVNMFQRASLIIFTLAFTIFVSAIVLGGEGISDMIGQIQRSEYMGFEDLC 212
A+ GQ ++ +L+ RASLIIF LA IF+SAI LGG GI +MIG+IQR EYMGFE+LC
Sbjct: 413 AWVGQHVVRRLIAAIGRASLIIFILASMIFISAISLGGVGIVNMIGKIQRHEYMGFENLC 472
Query: 211 KY 206
KY
Sbjct: 473 KY 474
>gb|AAL27555.1|AF420410_1 hypothetical protein [Musa acuminata]
Length = 238
Score = 151 bits (381), Expect = 6e-36
Identities = 73/115 (63%), Positives = 93/115 (80%)
Frame = -3
Query: 571 GGGFVMGPLFLELGIAPQVASATATFGMTFSASISVVQYYLLNRFPVPYALYLTLVAAIA 392
GGGF++GP+FLELG+ PQV+SATATF MTFS+S+SVV+YYLL RFP+PYALY VA +A
Sbjct: 83 GGGFILGPVFLELGVPPQVSSATATFAMTFSSSMSVVEYYLLKRFPIPYALYFVSVALVA 142
Query: 391 AYRGQFIIDKLVNMFQRASLIIFTLAFTIFVSAIVLGGEGISDMIGQIQRSEYMG 227
A+ GQ ++ +L+ + RASLIIF LA TIF+SAI LGG GIS+M+ +IQ E G
Sbjct: 143 AFVGQHLVKRLIEILGRASLIIFILASTIFISAISLGGVGISNMVQKIQHHESWG 197
>ref|NP_181201.2| unknown protein; protein id: At2g36630.1, supported by cDNA:
gi_18700128 [Arabidopsis thaliana]
gi|18700129|gb|AAL77676.1| At2g36630/F1O11.26
[Arabidopsis thaliana] gi|22137236|gb|AAM91463.1|
At2g36630/F1O11.26 [Arabidopsis thaliana]
Length = 459
Score = 133 bits (334), Expect = 2e-30
Identities = 66/120 (55%), Positives = 86/120 (71%)
Frame = -3
Query: 571 GGGFVMGPLFLELGIAPQVASATATFGMTFSASISVVQYYLLNRFPVPYALYLTLVAAIA 392
GGGFV+GPL LE+G+ PQVASATATF M FS+S+SVV++YLL RFP+PYA+YL V+ +A
Sbjct: 337 GGGFVLGPLLLEIGVIPQVASATATFVMMFSSSLSVVEFYLLKRFPIPYAMYLISVSILA 396
Query: 391 AYRGQFIIDKLVNMFQRASLIIFTLAFTIFVSAIVLGGEGISDMIGQIQRSEYMGFEDLC 212
+ GQ I KLV + +RAS+I+F L+ I SA+ +G GI I I E+MGF C
Sbjct: 397 GFWGQSFIRKLVAILRRASIIVFVLSGVICASALTMGVIGIEKSIKMIHNHEFMGFLGFC 456
>gb|AAF16643.1|AC011661_21 T23J18.20 [Arabidopsis thaliana]
Length = 491
Score = 82.0 bits (201), Expect = 5e-15
Identities = 40/120 (33%), Positives = 69/120 (57%)
Frame = -3
Query: 571 GGGFVMGPLFLELGIAPQVASATATFGMTFSASISVVQYYLLNRFPVPYALYLTLVAAIA 392
GGG ++ PL L++GIAP+V +AT +F + FS+S+S +QY LL A LV +A
Sbjct: 372 GGGMLISPLLLQIGIAPEVTAATCSFMVLFSSSMSAIQYLLLGMEHAGTAAIFALVCFVA 431
Query: 391 AYRGQFIIDKLVNMFQRASLIIFTLAFTIFVSAIVLGGEGISDMIGQIQRSEYMGFEDLC 212
+ G ++ K++ + RAS+I+F + + +S +++ G ++ YMGF+ C
Sbjct: 432 SLVGLMVVKKVIAKYGRASIIVFAVGIVMALSTVLMTTHGAFNVWNDFVSGRYMGFKLPC 491
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 484,539,688
Number of Sequences: 1393205
Number of extensions: 10228569
Number of successful extensions: 23257
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 22524
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23248
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)