Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002146A_C01 KMC002146A_c01
(645 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
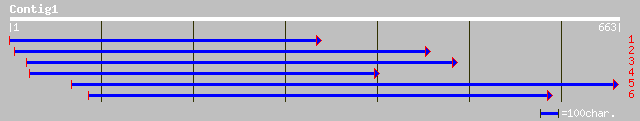
ref|NP_060875.1| hypothetical protein, clone 2746033 [Homo sapie... 33 2.5
gb|AAK97661.1|AF407672_1 TTP1 [Homo sapiens] 33 2.5
ref|NP_282107.1| phosphoribosylformylglycinamidine synthase [Cam... 32 7.3
sp|Q07891|TFR1_CRIGR Transferrin receptor protein 1 (TfR1) (TR) ... 32 9.5
emb|CAB52257.1| phosphoprotein phosphatase [Wolbachia sp. wRi] 32 9.5
>ref|NP_060875.1| hypothetical protein, clone 2746033 [Homo sapiens]
gi|15214530|gb|AAH12386.1|AAH12386 Unknown (protein for
MGC:17778) [Homo sapiens] gi|21912398|emb|CAB77267.2|
hypothetical protein [Homo sapiens]
Length = 184
Score = 33.5 bits (75), Expect = 2.5
Identities = 20/72 (27%), Positives = 30/72 (40%)
Frame = -2
Query: 533 TPMRSSNVYNYGEEFKLMEEPTINTGDNNTQAWEIPAATSTTNMEMPNYWSWEDIDSLVS 354
+P R ++ E F + EE + D WE+ T+ E P ED DS +
Sbjct: 66 SPARGEGTHSEEEGFAMDEEDS----DGELNTWELSEGTNCPPKEQPGDLFNEDWDSELK 121
Query: 353 TDLNVPWDDSDV 318
D P+D D+
Sbjct: 122 ADQGNPYDADDI 133
>gb|AAK97661.1|AF407672_1 TTP1 [Homo sapiens]
Length = 184
Score = 33.5 bits (75), Expect = 2.5
Identities = 20/72 (27%), Positives = 30/72 (40%)
Frame = -2
Query: 533 TPMRSSNVYNYGEEFKLMEEPTINTGDNNTQAWEIPAATSTTNMEMPNYWSWEDIDSLVS 354
+P R ++ E F + EE + D WE+ T+ E P ED DS +
Sbjct: 66 SPARGEGTHSEEEGFAMDEEDS----DGELNTWELSEGTNCPPKEQPGDLFNEDWDSELK 121
Query: 353 TDLNVPWDDSDV 318
D P+D D+
Sbjct: 122 ADQGNPYDADDI 133
>ref|NP_282107.1| phosphoribosylformylglycinamidine synthase [Campylobacter jejuni]
gi|14916672|sp|Q9PNY0|PURL_CAMJE
Phosphoribosylformylglycinamidine synthase II (FGAM
synthase II) gi|11272639|pir||B81370
phosphoribosylformylglycinamidine synthase (EC 6.3.5.3)
Cj0955c [imported] - Campylobacter jejuni (strain NCTC
11168) gi|6968392|emb|CAB73212.1|
phosphoribosylformylglycinamidine synthase
[Campylobacter jejuni subsp. jejuni NCTC 11168]
Length = 728
Score = 32.0 bits (71), Expect = 7.3
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 2/50 (4%)
Frame = -2
Query: 608 PQNQSFLFPFSTSCSLDTNPCAAVSTPMRSSNV--YNYGEEFKLMEEPTI 465
PQN ++ F+ C C ++TP+ S NV YN E + PTI
Sbjct: 498 PQNPEVMWQFAQGCEGIKEACKELNTPVVSGNVSLYNETEGVSIYPSPTI 547
>sp|Q07891|TFR1_CRIGR Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr)
gi|539790|pir||A48592 transferrin receptor protein -
Chinese hamster gi|304529|gb|AAA03576.1| transferrin
receptor
Length = 757
Score = 31.6 bits (70), Expect = 9.5
Identities = 19/50 (38%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Frame = +3
Query: 171 NSH--MKIFISKQENSLTNLYYETQKHPQLNNY*SMHTTIMVMCFFFSGF 314
NSH MK+ + ++EN+ N+ +KH +LN TI V+ FF GF
Sbjct: 33 NSHVEMKLAVDEEENTDNNMKASVRKHRRLNGR-LCFGTIAVVIFFLIGF 81
>emb|CAB52257.1| phosphoprotein phosphatase [Wolbachia sp. wRi]
Length = 428
Score = 31.6 bits (70), Expect = 9.5
Identities = 14/39 (35%), Positives = 22/39 (55%)
Frame = -2
Query: 512 VYNYGEEFKLMEEPTINTGDNNTQAWEIPAATSTTNMEM 396
VY GEE +++ +IN GD+N E+ TS N+ +
Sbjct: 52 VYVDGEETEMLNRSSINNGDHNETVCELTVQTSAKNISI 90
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 551,214,770
Number of Sequences: 1393205
Number of extensions: 11905920
Number of successful extensions: 28335
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 27492
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28320
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27291941472
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)