Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
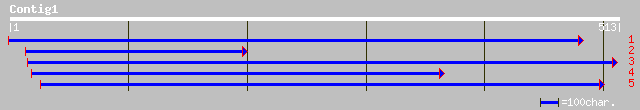
Query= KMC002145A_C01 KMC002145A_c01
(510 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA72330.1| shaggy-like kinase [Ricinus communis] 167 8e-41
gb|AAM65084.1| putative shaggy-like protein kinase dzeta [Arabid... 165 3e-40
sp|Q39010|KSGD_ARATH Shaggy-related protein kinase delta (ASK-de... 165 3e-40
ref|NP_180655.1| putative shaggy-like protein kinase dzeta; prot... 165 3e-40
sp|Q39011|KSGE_ARATH Shaggy-related protein kinase eta (ASK-eta)... 164 5e-40
>emb|CAA72330.1| shaggy-like kinase [Ricinus communis]
Length = 277
Score = 167 bits (422), Expect = 8e-41
Identities = 78/92 (84%), Positives = 86/92 (92%)
Frame = -3
Query: 508 FHKRMPPEAIDLASRLLQYSPSLRCSALEACAHPFFDELREPNARLPNGRPLPPLFNFRQ 329
FHKRMPPEAIDLASRLLQYSPSLRC+ALEACAHPFFDELREPNARLPNGRPLPPLFNF+Q
Sbjct: 186 FHKRMPPEAIDLASRLLQYSPSLRCTALEACAHPFFDELREPNARLPNGRPLPPLFNFKQ 245
Query: 328 ELGGASPELIIKLIPEHVRRQTGLSFPYPAST 233
EL GASPEL+ KLIP+H++RQ GL+F + A T
Sbjct: 246 ELNGASPELVNKLIPDHMKRQMGLNFLHLAGT 277
>gb|AAM65084.1| putative shaggy-like protein kinase dzeta [Arabidopsis thaliana]
Length = 412
Score = 165 bits (417), Expect = 3e-40
Identities = 78/90 (86%), Positives = 83/90 (91%)
Frame = -3
Query: 508 FHKRMPPEAIDLASRLLQYSPSLRCSALEACAHPFFDELREPNARLPNGRPLPPLFNFRQ 329
FHKRMPPEAIDLASRLLQYSPSLRC+ALEACAHPFF+ELREPNARLPNGRPLPPLFNF+Q
Sbjct: 321 FHKRMPPEAIDLASRLLQYSPSLRCTALEACAHPFFNELREPNARLPNGRPLPPLFNFKQ 380
Query: 328 ELGGASPELIIKLIPEHVRRQTGLSFPYPA 239
EL GASPELI +LIPEHVRRQ FP+ A
Sbjct: 381 ELSGASPELINRLIPEHVRRQMNGGFPFQA 410
>sp|Q39010|KSGD_ARATH Shaggy-related protein kinase delta (ASK-delta) (ASK-dzeta)
gi|2129738|pir||S71266 shaggy-like protein kinase zeta
(EC 2.7.1.-) - Arabidopsis thaliana
gi|1225913|emb|CAA64408.1| shaggy-like kinase dzeta
[Arabidopsis thaliana] gi|1669653|emb|CAA70483.1|
serine/threonine kinase [Arabidopsis thaliana]
Length = 412
Score = 165 bits (417), Expect = 3e-40
Identities = 78/90 (86%), Positives = 83/90 (91%)
Frame = -3
Query: 508 FHKRMPPEAIDLASRLLQYSPSLRCSALEACAHPFFDELREPNARLPNGRPLPPLFNFRQ 329
FHKRMPPEAIDLASRLLQYSPSLRC+ALEACAHPFF+ELREPNARLPNGRPLPPLFNF+Q
Sbjct: 321 FHKRMPPEAIDLASRLLQYSPSLRCTALEACAHPFFNELREPNARLPNGRPLPPLFNFKQ 380
Query: 328 ELGGASPELIIKLIPEHVRRQTGLSFPYPA 239
EL GASPELI +LIPEHVRRQ FP+ A
Sbjct: 381 ELSGASPELINRLIPEHVRRQMNGGFPFQA 410
>ref|NP_180655.1| putative shaggy-like protein kinase dzeta; protein id: At2g30980.1,
supported by cDNA: 36479., supported by cDNA:
gi_17381127, supported by cDNA: gi_20453107, supported
by cDNA: gi_20465726 [Arabidopsis thaliana]
gi|25287637|pir||A84715 probable shaggy-like protein
kinase dzeta [imported] - Arabidopsis thaliana
gi|3201623|gb|AAC20732.1| putative shaggy-like protein
kinase dzeta [Arabidopsis thaliana]
gi|17381128|gb|AAL36376.1| putative shaggy protein
kinase dzeta [Arabidopsis thaliana]
gi|20453108|gb|AAM19796.1| At2g30980/F7F1.19
[Arabidopsis thaliana] gi|20465727|gb|AAM20332.1|
putative shaggy protein kinase dzeta [Arabidopsis
thaliana]
Length = 412
Score = 165 bits (417), Expect = 3e-40
Identities = 78/90 (86%), Positives = 83/90 (91%)
Frame = -3
Query: 508 FHKRMPPEAIDLASRLLQYSPSLRCSALEACAHPFFDELREPNARLPNGRPLPPLFNFRQ 329
FHKRMPPEAIDLASRLLQYSPSLRC+ALEACAHPFF+ELREPNARLPNGRPLPPLFNF+Q
Sbjct: 321 FHKRMPPEAIDLASRLLQYSPSLRCTALEACAHPFFNELREPNARLPNGRPLPPLFNFKQ 380
Query: 328 ELGGASPELIIKLIPEHVRRQTGLSFPYPA 239
EL GASPELI +LIPEHVRRQ FP+ A
Sbjct: 381 ELSGASPELINRLIPEHVRRQMNGGFPFQA 410
>sp|Q39011|KSGE_ARATH Shaggy-related protein kinase eta (ASK-eta)
gi|1161512|emb|CAA64409.1| shaggy-like kinase etha
[Arabidopsis thaliana] gi|1627516|emb|CAA70144.1|
shaggy-like kinase etha [Arabidopsis thaliana]
Length = 380
Score = 164 bits (415), Expect = 5e-40
Identities = 74/86 (86%), Positives = 83/86 (96%)
Frame = -3
Query: 508 FHKRMPPEAIDLASRLLQYSPSLRCSALEACAHPFFDELREPNARLPNGRPLPPLFNFRQ 329
FHKRMPPEA+DLASRLLQYSPSLRC+ALEACAHPFFDELREPNARLPNGRPLPPLFNF+Q
Sbjct: 289 FHKRMPPEAVDLASRLLQYSPSLRCTALEACAHPFFDELREPNARLPNGRPLPPLFNFKQ 348
Query: 328 ELGGASPELIIKLIPEHVRRQTGLSF 251
E+ G+SPEL+ KLIP+H++RQ GLSF
Sbjct: 349 EVAGSSPELVNKLIPDHIKRQLGLSF 374
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 437,850,304
Number of Sequences: 1393205
Number of extensions: 9496367
Number of successful extensions: 25091
Number of sequences better than 10.0: 740
Number of HSP's better than 10.0 without gapping: 24310
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25024
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 15942513235
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)