Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002142A_C01 KMC002142A_c01
(629 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
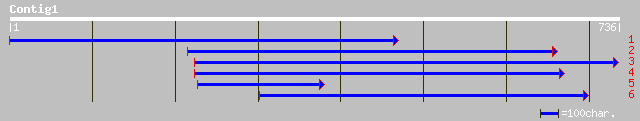
gb|AAO72531.1| alcohol dehydrogenase 1; ADH1 [Lotus corniculatus] 164 5e-73
gb|AAC97495.1| alcohol-dehydrogenase [Glycine max] 156 3e-70
sp|P12886|ADH1_PEA Alcohol dehydrogenase 1 gi|81891|pir||S00912 ... 156 5e-70
sp|P13603|ADH1_TRIRP Alcohol dehydrogenase 1 gi|65900|pir||DEJYA... 155 1e-69
pir||S53307 alcohol dehydrogenase (EC 1.1.1.1) 1 - Phaseolus acu... 154 6e-69
>gb|AAO72531.1| alcohol dehydrogenase 1; ADH1 [Lotus corniculatus]
Length = 380
Score = 164 bits (414), Expect(2) = 5e-73
Identities = 76/77 (98%), Positives = 76/77 (98%)
Frame = +1
Query: 397 GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINTDRGVMISDNQSR 576
GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINTDRGVMISDNQSR
Sbjct: 74 GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINTDRGVMISDNQSR 133
Query: 577 FSIKGKPIYHLVGTSTF 627
FSIKGKPIYH VGTSTF
Sbjct: 134 FSIKGKPIYHFVGTSTF 150
Score = 132 bits (333), Expect(2) = 5e-73
Identities = 62/62 (100%), Positives = 62/62 (100%)
Frame = +3
Query: 213 AAVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEA 392
AAVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEA
Sbjct: 13 AAVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEA 72
Query: 393 GG 398
GG
Sbjct: 73 GG 74
>gb|AAC97495.1| alcohol-dehydrogenase [Glycine max]
Length = 341
Score = 156 bits (394), Expect(2) = 3e-70
Identities = 71/77 (92%), Positives = 75/77 (97%)
Frame = +1
Query: 397 GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINTDRGVMISDNQSR 576
GIVESVGEGVTHLKPGDHALPVFTGECG+CPHCKSEESNMCDLLRINTDRGVMI D+Q+R
Sbjct: 73 GIVESVGEGVTHLKPGDHALPVFTGECGDCPHCKSEESNMCDLLRINTDRGVMIHDSQTR 132
Query: 577 FSIKGKPIYHLVGTSTF 627
FSIKG+PIYH VGTSTF
Sbjct: 133 FSIKGQPIYHFVGTSTF 149
Score = 131 bits (329), Expect(2) = 3e-70
Identities = 61/61 (100%), Positives = 61/61 (100%)
Frame = +3
Query: 216 AVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAG 395
AVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAG
Sbjct: 13 AVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAG 72
Query: 396 G 398
G
Sbjct: 73 G 73
>sp|P12886|ADH1_PEA Alcohol dehydrogenase 1 gi|81891|pir||S00912 alcohol dehydrogenase
(EC 1.1.1.1) 1 - garden pea gi|20639|emb|CAA29609.1|
alcohol dehydrogenase [Pisum sativum]
Length = 380
Score = 156 bits (395), Expect(2) = 5e-70
Identities = 70/77 (90%), Positives = 76/77 (97%)
Frame = +1
Query: 397 GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINTDRGVMISDNQSR 576
GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINTDRGVM++DN+SR
Sbjct: 74 GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINTDRGVMLNDNKSR 133
Query: 577 FSIKGKPIYHLVGTSTF 627
FSIKG+P++H VGTSTF
Sbjct: 134 FSIKGQPVHHFVGTSTF 150
Score = 130 bits (326), Expect(2) = 5e-70
Identities = 60/62 (96%), Positives = 62/62 (99%)
Frame = +3
Query: 213 AAVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEA 392
AAV+WEAGKPLVIEEVEVAPPQAGEVRLKIL+TSLCHTDVYFWEAKGQTPLFPRIFGHEA
Sbjct: 13 AAVAWEAGKPLVIEEVEVAPPQAGEVRLKILFTSLCHTDVYFWEAKGQTPLFPRIFGHEA 72
Query: 393 GG 398
GG
Sbjct: 73 GG 74
>sp|P13603|ADH1_TRIRP Alcohol dehydrogenase 1 gi|65900|pir||DEJYAW alcohol dehydrogenase
(EC 1.1.1.1) 1 - white clover gi|21951|emb|CAA32934.1|
alcohol dehydrogenase I (AA 1-380) [Trifolium repens]
Length = 380
Score = 155 bits (392), Expect(2) = 1e-69
Identities = 70/77 (90%), Positives = 76/77 (97%)
Frame = +1
Query: 397 GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINTDRGVMISDNQSR 576
GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMC+LLRINTDRGVMI+DN+SR
Sbjct: 74 GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCNLLRINTDRGVMINDNKSR 133
Query: 577 FSIKGKPIYHLVGTSTF 627
FSIKG+P++H VGTSTF
Sbjct: 134 FSIKGQPVHHFVGTSTF 150
Score = 130 bits (326), Expect(2) = 1e-69
Identities = 60/62 (96%), Positives = 62/62 (99%)
Frame = +3
Query: 213 AAVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEA 392
AAV+WEAGKPLVIEEVEVAPPQAGEVRLKIL+TSLCHTDVYFWEAKGQTPLFPRIFGHEA
Sbjct: 13 AAVAWEAGKPLVIEEVEVAPPQAGEVRLKILFTSLCHTDVYFWEAKGQTPLFPRIFGHEA 72
Query: 393 GG 398
GG
Sbjct: 73 GG 74
>pir||S53307 alcohol dehydrogenase (EC 1.1.1.1) 1 - Phaseolus acutifolius
gi|452769|emb|CAA80691.1| alcohol dehydrogenase-1F
[Phaseolus acutifolius]
Length = 380
Score = 154 bits (389), Expect(2) = 6e-69
Identities = 71/77 (92%), Positives = 74/77 (95%)
Frame = +1
Query: 397 GIVESVGEGVTHLKPGDHALPVFTGECGECPHCKSEESNMCDLLRINTDRGVMISDNQSR 576
GIVESVGEGVTHLKPGDHALPVFTGECGEC HCKSEESNMCDLLRINTDRGVMI D+Q+R
Sbjct: 74 GIVESVGEGVTHLKPGDHALPVFTGECGECAHCKSEESNMCDLLRINTDRGVMIHDSQTR 133
Query: 577 FSIKGKPIYHLVGTSTF 627
FSIKG+PIYH VGTSTF
Sbjct: 134 FSIKGQPIYHFVGTSTF 150
Score = 129 bits (323), Expect(2) = 6e-69
Identities = 59/62 (95%), Positives = 62/62 (99%)
Frame = +3
Query: 213 AAVSWEAGKPLVIEEVEVAPPQAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEA 392
AAV+WEAGKPLV+EEVEVAPP+AGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEA
Sbjct: 13 AAVAWEAGKPLVMEEVEVAPPKAGEVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEA 72
Query: 393 GG 398
GG
Sbjct: 73 GG 74
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 584,047,737
Number of Sequences: 1393205
Number of extensions: 13435131
Number of successful extensions: 45087
Number of sequences better than 10.0: 1380
Number of HSP's better than 10.0 without gapping: 41032
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44614
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)