Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002139A_C01 KMC002139A_c01
(766 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
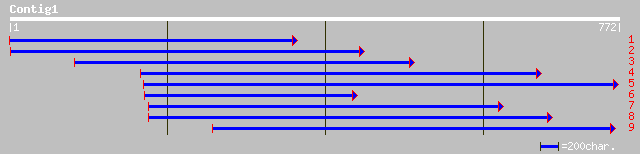
ref|NP_704401.1| hypothetical protein [Plasmodium falciparum 3D7... 38 0.18
gb|AAM98086.1| At1g73430/T9L24_16 [Arabidopsis thaliana] 36 0.69
ref|NP_493026.1| Histone DeAcetylase HDA-3 (52.7 kD) (hda-3) [Ca... 33 5.8
gb|EAA20719.1| hypothetical protein [Plasmodium yoelii yoelii] 32 7.6
ref|NP_703497.1| phosphatidylinositol 3-kinase, putative [Plasmo... 32 7.6
>ref|NP_704401.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23499140|emb|CAD51220.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 1182
Score = 37.7 bits (86), Expect = 0.18
Identities = 26/106 (24%), Positives = 48/106 (44%), Gaps = 1/106 (0%)
Frame = +2
Query: 422 SEKAISVKQSLIDLYTWSCQNLSNYKLDSAYNGLLSSFNIPEELP-SLVNILYE*KKLPA 598
+EK ++ + I Y + ++N+ + A N NIP L + NIL +
Sbjct: 601 TEKEKNMIYNYIHSYNIERKLVANFTISQAVNNYEKGKNIPYILGYNQDNILID------ 654
Query: 599 *NAVPIIQLKGYNGMNLRNHRSKNIKLKNYRDHNNSEKQCETYVYN 736
+ ++ + N N+ + N N +++NN+ K C TY+YN
Sbjct: 655 ----KLSKMCNFKSTNCHNNNNNNNNNNNNKNNNNNNKSCGTYIYN 696
>gb|AAM98086.1| At1g73430/T9L24_16 [Arabidopsis thaliana]
Length = 784
Score = 35.8 bits (81), Expect = 0.69
Identities = 24/49 (48%), Positives = 27/49 (54%), Gaps = 8/49 (16%)
Frame = -2
Query: 591 SFFYSYNMFTSEGNSSGMLK--------LESNPLYAESSL*LLKFWQLQ 469
S FYS NM S N +LK +E NP YAESS+ LLKF Q Q
Sbjct: 194 SNFYSPNMNVSNSNFLPLLKRLDECISYIEDNPQYAESSVYLLKFRQPQ 242
>ref|NP_493026.1| Histone DeAcetylase HDA-3 (52.7 kD) (hda-3) [Caenorhabditis
elegans] gi|7506349|pir||T23963 hypothetical protein
R06C1.1 - Caenorhabditis elegans
gi|3878853|emb|CAB03224.1| Hypothetical protein R06C1.1
[Caenorhabditis elegans] gi|3879019|emb|CAB03240.1|
Hypothetical protein R06C1.1 [Caenorhabditis elegans]
Length = 465
Score = 32.7 bits (73), Expect = 5.8
Identities = 29/116 (25%), Positives = 56/116 (48%), Gaps = 15/116 (12%)
Frame = +2
Query: 425 EKAISVKQSLID---LYTWSCQNLSNYKLDSAYNGLLSSFNIPEEL-PSLVNILYE*KKL 592
E AI++ Q + D L+ + + +YKL LS+FN PE + ++V +L K+L
Sbjct: 314 ETAIALNQEVSDDLPLHDYFDYFIPDYKLHIKPLAALSNFNTPEFIDQTIVALLENLKQL 373
Query: 593 PA*NAVPIIQL-----------KGYNGMNLRNHRSKNIKLKNYRDHNNSEKQCETY 727
P VP +Q+ K ++ +R+H++ ++++ + + E E Y
Sbjct: 374 P---HVPSVQMQSISTSCDSIVKTFDEKLIRDHQNDDVRVTQFEEDVQVEDSAEFY 426
>gb|EAA20719.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 869
Score = 32.3 bits (72), Expect = 7.6
Identities = 25/110 (22%), Positives = 52/110 (46%)
Frame = +2
Query: 407 KLYFRSEKAISVKQSLIDLYTWSCQNLSNYKLDSAYNGLLSSFNIPEELPSLVNILYE*K 586
K+ ++S I +K+ L +C+N+ ++ ++ +I ++ N +Y +
Sbjct: 497 KIKYQSGNTIEIKEKEAKL---NCENMHLMDKENRTYSIIKIASIISKIDKSKNSIYN-E 552
Query: 587 KLPA*NAVPIIQLKGYNGMNLRNHRSKNIKLKNYRDHNNSEKQCETYVYN 736
P N I +++G+ + N K+I +K+Y +N S K C Y+ N
Sbjct: 553 NEPNENKTNI-KIEGF----IFNEHLKDITIKSYNINNKSSKNCIKYISN 597
>ref|NP_703497.1| phosphatidylinositol 3-kinase, putative [Plasmodium falciparum 3D7]
gi|23504638|emb|CAD51517.1| phosphatidylinositol
3-kinase, putative [Plasmodium falciparum 3D7]
Length = 2133
Score = 32.3 bits (72), Expect = 7.6
Identities = 19/61 (31%), Positives = 27/61 (44%), Gaps = 14/61 (22%)
Frame = +2
Query: 623 LKGYNGMNLRNHRSKNIKLKNYRDHNNSEKQCETYVY--------------NIIGISSFI 760
L+G G N + ++KN K Y DH N + + Y Y N+ ISSF+
Sbjct: 92 LEGNQGFNKKPEKNKNTKGNVYTDHTNQNAKSKIYNYDMNDDSYSNYVNNNNVFRISSFL 151
Query: 761 I 763
I
Sbjct: 152 I 152
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 602,339,283
Number of Sequences: 1393205
Number of extensions: 12414700
Number of successful extensions: 24660
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 23285
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24638
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 37254821786
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)