Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002135A_C01 KMC002135A_c01
(523 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
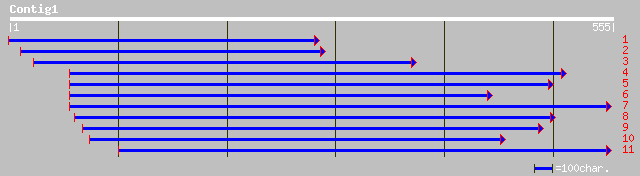
ref|NP_564766.1| putative iron-regulated transporter; protein id... 129 3e-51
pir||B96635 hypothetical protein T7P1.10 [imported] - Arabidopsi... 129 3e-51
gb|AAM62825.1| putative iron-regulated transporter [Arabidopsis ... 129 3e-51
ref|NP_172566.1| ZIP4, a putative zinc transporter; protein id: ... 128 4e-51
gb|AAK69430.1|AF275752_1 ZIP-like zinc transporter [Thlaspi caer... 127 6e-51
>ref|NP_564766.1| putative iron-regulated transporter; protein id: At1g60960.1,
supported by cDNA: 15980. [Arabidopsis thaliana]
gi|17385796|gb|AAL38438.1|AF369915_1 putative metal
transporter IRT3 [Arabidopsis thaliana]
Length = 425
Score = 129 bits (324), Expect(3) = 3e-51
Identities = 61/72 (84%), Positives = 66/72 (90%)
Frame = -3
Query: 521 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKTSSATIMACFFALTTPVGIGIGTGIA 342
QSPCTIRPLIAALSFHQFFEGFALGGCISQAQF+ SATIMACFFALTTP+GIGIGT +A
Sbjct: 295 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFRNKSATIMACFFALTTPIGIGIGTAVA 354
Query: 341 SVYNPYSAGALV 306
S +N +S GALV
Sbjct: 355 SSFNSHSVGALV 366
Score = 59.3 bits (142), Expect(3) = 3e-51
Identities = 28/34 (82%), Positives = 30/34 (87%)
Frame = -2
Query: 228 FSAKRMSCNFRLQIVSYCMLFLGAGLMSSLAIWA 127
F + +M CNFRLQIVSY MLFLGAGLMSSLAIWA
Sbjct: 392 FLSTKMRCNFRLQIVSYVMLFLGAGLMSSLAIWA 425
Score = 55.8 bits (133), Expect(3) = 3e-51
Identities = 29/36 (80%), Positives = 31/36 (85%)
Frame = -1
Query: 322 ALAPLYAEGILDSLSAGILVYMALVDLIAADFLSQK 215
++ L EGILDSLSAGILVYMALVDLIAADFLS K
Sbjct: 361 SVGALVTEGILDSLSAGILVYMALVDLIAADFLSTK 396
>pir||B96635 hypothetical protein T7P1.10 [imported] - Arabidopsis thaliana
gi|12323339|gb|AAG51647.1|AC018908_13 putative
iron-regulated transporter; 41763-40495 [Arabidopsis
thaliana]
Length = 389
Score = 129 bits (324), Expect(3) = 3e-51
Identities = 61/72 (84%), Positives = 66/72 (90%)
Frame = -3
Query: 521 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKTSSATIMACFFALTTPVGIGIGTGIA 342
QSPCTIRPLIAALSFHQFFEGFALGGCISQAQF+ SATIMACFFALTTP+GIGIGT +A
Sbjct: 259 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFRNKSATIMACFFALTTPIGIGIGTAVA 318
Query: 341 SVYNPYSAGALV 306
S +N +S GALV
Sbjct: 319 SSFNSHSVGALV 330
Score = 59.3 bits (142), Expect(3) = 3e-51
Identities = 28/34 (82%), Positives = 30/34 (87%)
Frame = -2
Query: 228 FSAKRMSCNFRLQIVSYCMLFLGAGLMSSLAIWA 127
F + +M CNFRLQIVSY MLFLGAGLMSSLAIWA
Sbjct: 356 FLSTKMRCNFRLQIVSYVMLFLGAGLMSSLAIWA 389
Score = 55.8 bits (133), Expect(3) = 3e-51
Identities = 29/36 (80%), Positives = 31/36 (85%)
Frame = -1
Query: 322 ALAPLYAEGILDSLSAGILVYMALVDLIAADFLSQK 215
++ L EGILDSLSAGILVYMALVDLIAADFLS K
Sbjct: 325 SVGALVTEGILDSLSAGILVYMALVDLIAADFLSTK 360
>gb|AAM62825.1| putative iron-regulated transporter [Arabidopsis thaliana]
Length = 389
Score = 129 bits (324), Expect(3) = 3e-51
Identities = 61/72 (84%), Positives = 66/72 (90%)
Frame = -3
Query: 521 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKTSSATIMACFFALTTPVGIGIGTGIA 342
QSPCTIRPLIAALSFHQFFEGFALGGCISQAQF+ SATIMACFFALTTP+GIGIGT +A
Sbjct: 259 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFRNKSATIMACFFALTTPIGIGIGTAVA 318
Query: 341 SVYNPYSAGALV 306
S +N +S GALV
Sbjct: 319 SSFNSHSVGALV 330
Score = 59.3 bits (142), Expect(3) = 3e-51
Identities = 28/34 (82%), Positives = 30/34 (87%)
Frame = -2
Query: 228 FSAKRMSCNFRLQIVSYCMLFLGAGLMSSLAIWA 127
F + +M CNFRLQIVSY MLFLGAGLMSSLAIWA
Sbjct: 356 FLSTKMRCNFRLQIVSYVMLFLGAGLMSSLAIWA 389
Score = 55.8 bits (133), Expect(3) = 3e-51
Identities = 29/36 (80%), Positives = 31/36 (85%)
Frame = -1
Query: 322 ALAPLYAEGILDSLSAGILVYMALVDLIAADFLSQK 215
++ L EGILDSLSAGILVYMALVDLIAADFLS K
Sbjct: 325 SVGALVTEGILDSLSAGILVYMALVDLIAADFLSTK 360
>ref|NP_172566.1| ZIP4, a putative zinc transporter; protein id: At1g10970.1
[Arabidopsis thaliana] gi|25347103|pir||F86243 ZIP4,
probable zinc transporter, 61460-62785 [imported] -
Arabidopsis thaliana gi|1931645|gb|AAB65480.1| ZIP4, a
putative zinc transporter; 61460-62785 [Arabidopsis
thaliana]
Length = 374
Score = 128 bits (321), Expect(3) = 4e-51
Identities = 61/72 (84%), Positives = 66/72 (90%)
Frame = -3
Query: 521 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKTSSATIMACFFALTTPVGIGIGTGIA 342
QSPCTIRPLIAALSFHQFFEGFALGGCISQAQF+ SATIMACFFALTTP+GIGIGT +A
Sbjct: 244 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFRNKSATIMACFFALTTPLGIGIGTAVA 303
Query: 341 SVYNPYSAGALV 306
S +N +S GALV
Sbjct: 304 SSFNSHSPGALV 315
Score = 60.8 bits (146), Expect(3) = 4e-51
Identities = 28/34 (82%), Positives = 31/34 (90%)
Frame = -2
Query: 228 FSAKRMSCNFRLQIVSYCMLFLGAGLMSSLAIWA 127
F +KRMSCN RLQ+VSY MLFLGAGLMS+LAIWA
Sbjct: 341 FLSKRMSCNLRLQVVSYVMLFLGAGLMSALAIWA 374
Score = 55.1 bits (131), Expect(3) = 4e-51
Identities = 28/32 (87%), Positives = 30/32 (93%)
Frame = -1
Query: 310 LYAEGILDSLSAGILVYMALVDLIAADFLSQK 215
L EGILDSLSAGILVYMALVDLIAADFLS++
Sbjct: 314 LVTEGILDSLSAGILVYMALVDLIAADFLSKR 345
>gb|AAK69430.1|AF275752_1 ZIP-like zinc transporter [Thlaspi caerulescens]
Length = 422
Score = 127 bits (318), Expect(3) = 6e-51
Identities = 61/72 (84%), Positives = 65/72 (89%)
Frame = -3
Query: 521 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKTSSATIMACFFALTTPVGIGIGTGIA 342
QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFK SATIMACFFALTTP+ IGIGT +A
Sbjct: 292 QSPCTIRPLIAALSFHQFFEGFALGGCISQAQFKNKSATIMACFFALTTPISIGIGTVVA 351
Query: 341 SVYNPYSAGALV 306
S +N +S GALV
Sbjct: 352 SSFNAHSVGALV 363
Score = 60.8 bits (146), Expect(3) = 6e-51
Identities = 28/33 (84%), Positives = 31/33 (93%)
Frame = -2
Query: 228 FSAKRMSCNFRLQIVSYCMLFLGAGLMSSLAIW 130
F +KRMSCNFRLQIVSY +LFLG+GLMSSLAIW
Sbjct: 389 FLSKRMSCNFRLQIVSYLLLFLGSGLMSSLAIW 421
Score = 55.5 bits (132), Expect(3) = 6e-51
Identities = 28/36 (77%), Positives = 32/36 (88%)
Frame = -1
Query: 322 ALAPLYAEGILDSLSAGILVYMALVDLIAADFLSQK 215
++ L EGILDSLSAGILVYMALVDLIAADFLS++
Sbjct: 358 SVGALVTEGILDSLSAGILVYMALVDLIAADFLSKR 393
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 445,489,412
Number of Sequences: 1393205
Number of extensions: 9367854
Number of successful extensions: 25757
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 24934
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25721
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16731298976
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)