Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002131A_C01 KMC002131A_c01
(660 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
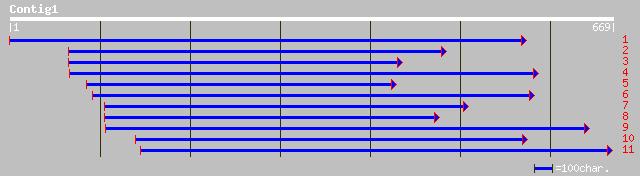
ref|NP_564234.1| expressed protein; protein id: At1g25400.1, sup... 69 4e-11
ref|NP_564930.1| expressed protein; protein id: At1g68440.1, sup... 59 6e-08
gb|AAG52388.1|AC011915_2 unknown protein, 5' partial; 67-381 [Ar... 57 2e-07
ref|XP_039353.1| hypothetical protein DKFZp547L112 [Homo sapiens] 49 6e-05
ref|NP_509491.1| Putative nuclear protein, nematode specific (11... 46 4e-04
>ref|NP_564234.1| expressed protein; protein id: At1g25400.1, supported by cDNA:
103226. [Arabidopsis thaliana] gi|25372926|pir||A86384
unknown protein [imported] - Arabidopsis thaliana
gi|12321508|gb|AAG50811.1|AC079281_13 unknown protein
[Arabidopsis thaliana] gi|21536564|gb|AAM60896.1|
unknown [Arabidopsis thaliana]
Length = 288
Score = 69.3 bits (168), Expect = 4e-11
Identities = 38/93 (40%), Positives = 56/93 (59%), Gaps = 3/93 (3%)
Frame = -3
Query: 592 SSSTASPAVVVSAAEGSRGNLEVGIWDTRLRKRRPSVVAEW---GPAVGKTVRVESGGVS 422
S S S AV+++A + LEV WD R+ P+++AEW G +GK +RV+ G V
Sbjct: 186 SYSLLSSAVLLAAEKKGSDGLEVSAWDARVGFGVPALLAEWKQPGRLLGKIIRVDVGDVD 245
Query: 421 KVYVRDNGRDGVTVGDMRKVSKPLGNVTESDAD 323
K+YV D+ +TVGDMR V+ L +TES+ +
Sbjct: 246 KIYVGDDVEGEITVGDMRMVNGALTELTESEVE 278
>ref|NP_564930.1| expressed protein; protein id: At1g68440.1, supported by cDNA:
34166., supported by cDNA: gi_16648772 [Arabidopsis
thaliana] gi|25372927|pir||D96708 unknown protein,
9003-8083 [imported] - Arabidopsis thaliana
gi|6714347|gb|AAF26038.1|AC015986_1 unknown protein;
9003-8083 [Arabidopsis thaliana]
gi|16648773|gb|AAL25577.1| At1g68440/T2E12_1
[Arabidopsis thaliana] gi|21592921|gb|AAM64871.1|
unknown [Arabidopsis thaliana]
gi|21700823|gb|AAM70535.1| At1g68440/T2E12_1
[Arabidopsis thaliana]
Length = 306
Score = 58.9 bits (141), Expect = 6e-08
Identities = 39/111 (35%), Positives = 62/111 (55%), Gaps = 4/111 (3%)
Frame = -3
Query: 643 LGLDFDDYEES-PLHAPASSSTASPAVVVSAAEGSRGNLEVGIWDTRLRKRRPSVVAEW- 470
L +D DD+E +ST+SP + +G ++V D R R P+++AEW
Sbjct: 186 LDIDGDDHENVVATFLKNYNSTSSPFFWAAEKKGVDA-VKVKACDPRAGFRMPALLAEWR 244
Query: 469 --GPAVGKTVRVESGGVSKVYVRDNGRDGVTVGDMRKVSKPLGNVTESDAD 323
G +G + V++GGV KVYVRD+ + VGD+RK + L ++TE +A+
Sbjct: 245 QPGRLLGNIIGVDTGGVEKVYVRDDVSGEIAVGDLRKFNGVLTDLTECEAE 295
>gb|AAG52388.1|AC011915_2 unknown protein, 5' partial; 67-381 [Arabidopsis thaliana]
Length = 104
Score = 57.4 bits (137), Expect = 2e-07
Identities = 34/92 (36%), Positives = 55/92 (58%), Gaps = 3/92 (3%)
Frame = -3
Query: 589 SSTASPAVVVSAAEGSRGNLEVGIWDTRLRKRRPSVVAEW---GPAVGKTVRVESGGVSK 419
+ST+SP + +G ++V D R R P+++AEW G +G + V++GGV K
Sbjct: 3 NSTSSPFFWAAEKKGVDA-VKVKACDPRAGFRMPALLAEWRQPGRLLGNIIGVDTGGVEK 61
Query: 418 VYVRDNGRDGVTVGDMRKVSKPLGNVTESDAD 323
VYVRD+ + VGD+RK + L ++TE +A+
Sbjct: 62 VYVRDDVSGEIAVGDLRKFNGVLTDLTECEAE 93
>ref|XP_039353.1| hypothetical protein DKFZp547L112 [Homo sapiens]
Length = 129
Score = 48.9 bits (115), Expect = 6e-05
Identities = 29/78 (37%), Positives = 36/78 (45%), Gaps = 6/78 (7%)
Frame = +1
Query: 382 P*RRHVRYLSHKP------STRRQIPRARSSPRRAPTPPPPTVSASGGACPKSPPPSSRV 543
P +RH R H+P RAR S +R P PP P +A + PPSSR+
Sbjct: 50 PGQRHGRTRLHRPVGYHIGKPPAHPHRARDSDKREPLPPAPRTTA------RQEPPSSRI 103
Query: 544 SLPQLKQPPPGTPWKNSP 597
+P PGTPW SP
Sbjct: 104 LTMATVEPRPGTPWTKSP 121
>ref|NP_509491.1| Putative nuclear protein, nematode specific (112.0 kD)
[Caenorhabditis elegans] gi|7506276|pir||T28872
hypothetical protein R04E5.8 - Caenorhabditis elegans
gi|9803002|gb|AAG00010.1| Hypothetical protein R04E5.8
[Caenorhabditis elegans]
Length = 997
Score = 46.2 bits (108), Expect = 4e-04
Identities = 32/109 (29%), Positives = 40/109 (36%), Gaps = 13/109 (11%)
Frame = +1
Query: 313 PGYYPHQIPSRFLTAC*PFSCHQP*RRHVRYLSHKP-----------STRRQIPRARSSP 459
P YP+Q P HQP H +L H Q+P +
Sbjct: 70 PPPYPYQHP------------HQPPPAHPHHLQHPHPYAYPGYPVPGQEGHQVPAPQHGD 117
Query: 460 RRA--PTPPPPTVSASGGACPKSPPPSSRVSLPQLKQPPPGTPWKNSPA 600
A P PPPP S +GG+ P PPP P + PPP P P+
Sbjct: 118 HEASPPPPPPPRKSRAGGSSPPPPPPPRVPRTPPPRSPPPRRPPMTPPS 166
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 608,809,959
Number of Sequences: 1393205
Number of extensions: 15651177
Number of successful extensions: 131350
Number of sequences better than 10.0: 1739
Number of HSP's better than 10.0 without gapping: 74075
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 111329
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28289785200
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)