Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002127A_C01 KMC002127A_c01
(583 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
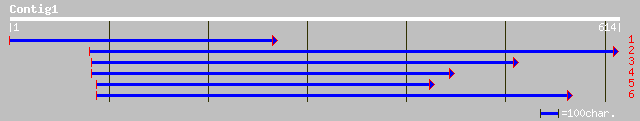
ref|NP_173320.1| WRKY family transcription factor; protein id: A... 66 3e-10
sp|Q8VWV6|WR61_ARATH Probable WRKY transcription factor 61 (WRKY... 66 3e-10
gb|AAL32032.1|AF439273_1 NADH-ubiquinone oxidoreductase [Retama ... 47 4e-09
ref|NP_568778.1| expressed protein; protein id: At5g52840.1, sup... 49 2e-07
ref|NP_192081.1| WRKY family transcription factor; protein id: A... 56 4e-07
>ref|NP_173320.1| WRKY family transcription factor; protein id: At1g18860.1
[Arabidopsis thaliana] gi|25518561|pir||E86322
hypothetical protein F6A14.5 - Arabidopsis thaliana
gi|6730700|gb|AAF27095.1|AC011809_4 Hypothetical protein
[Arabidopsis thaliana]
Length = 471
Score = 66.2 bits (160), Expect = 3e-10
Identities = 43/90 (47%), Positives = 54/90 (59%), Gaps = 3/90 (3%)
Frame = +1
Query: 202 GTHNHPLPVSATAMDSSTSAALSML*SGSYTSQPTGHCAASMGSASTMFIGLNFSSYNQS 381
GTHNHPLP+SATAM S+TSAA SML SG A+S SA+ GLNFS +
Sbjct: 235 GTHNHPLPMSATAMASATSAAASMLLSG----------ASSSSSAAADLHGLNFSLSGNN 284
Query: 382 ---RATQAFLPTPTPHLFPTITLDLT*TTS 462
+ FL +P+ PT+TLDLT ++S
Sbjct: 285 ITPKPKTHFLQSPSSSGHPTVTLDLTTSSS 314
>sp|Q8VWV6|WR61_ARATH Probable WRKY transcription factor 61 (WRKY DNA-binding protein 61)
gi|17980960|gb|AAL50785.1|AF452175_1 WRKY transcription
factor 61 [Arabidopsis thaliana]
Length = 480
Score = 66.2 bits (160), Expect = 3e-10
Identities = 43/90 (47%), Positives = 54/90 (59%), Gaps = 3/90 (3%)
Frame = +1
Query: 202 GTHNHPLPVSATAMDSSTSAALSML*SGSYTSQPTGHCAASMGSASTMFIGLNFSSYNQS 381
GTHNHPLP+SATAM S+TSAA SML SG A+S SA+ GLNFS +
Sbjct: 244 GTHNHPLPMSATAMASATSAAASMLLSG----------ASSSSSAAADLHGLNFSLSGNN 293
Query: 382 ---RATQAFLPTPTPHLFPTITLDLT*TTS 462
+ FL +P+ PT+TLDLT ++S
Sbjct: 294 ITPKPKTHFLQSPSSSGHPTVTLDLTTSSS 323
>gb|AAL32032.1|AF439273_1 NADH-ubiquinone oxidoreductase [Retama raetam]
Length = 153
Score = 46.6 bits (109), Expect(2) = 4e-09
Identities = 17/20 (85%), Positives = 18/20 (90%)
Frame = -1
Query: 574 PWGVPDDYECEAFDNDAPVP 515
PWGVPDDYECE +NDAPVP
Sbjct: 94 PWGVPDDYECEVIENDAPVP 113
Score = 35.8 bits (81), Expect(2) = 4e-09
Identities = 14/16 (87%), Positives = 14/16 (87%)
Frame = -3
Query: 515 KHVLLHRPPPLPKEFH 468
KHV LHRPPPLP EFH
Sbjct: 114 KHVPLHRPPPLPTEFH 129
>ref|NP_568778.1| expressed protein; protein id: At5g52840.1, supported by cDNA:
gi_15809965, supported by cDNA: gi_18958051 [Arabidopsis
thaliana] gi|25090853|sp|Q9FLX7|NUFM_ARATH Probable
NADH-ubiquinone oxidoreductase 18 kDa subunit,
mitochondrial precursor (Complex I-18Kd) (CI-18Kd)
gi|10177098|dbj|BAB10432.1| gene_id:MXC20.6~unknown
protein [Arabidopsis thaliana]
gi|15809966|gb|AAL06910.1| AT5g52840/MXC20_6
[Arabidopsis thaliana] gi|18958052|gb|AAL79599.1|
AT5g52840/MXC20_6 [Arabidopsis thaliana]
Length = 169
Score = 48.9 bits (115), Expect(2) = 2e-07
Identities = 18/23 (78%), Positives = 20/23 (86%)
Frame = -1
Query: 583 E*DPWGVPDDYECEAFDNDAPVP 515
E DPWGVPDDYECE +NDAP+P
Sbjct: 105 EWDPWGVPDDYECEVIENDAPIP 127
Score = 27.3 bits (59), Expect(2) = 2e-07
Identities = 10/16 (62%), Positives = 13/16 (80%)
Frame = -3
Query: 515 KHVLLHRPPPLPKEFH 468
KHV HRP PLP++F+
Sbjct: 128 KHVPQHRPGPLPEQFY 143
>ref|NP_192081.1| WRKY family transcription factor; protein id: At4g01720.1,
supported by cDNA: gi_19172391 [Arabidopsis thaliana]
gi|20978798|sp|Q9ZSI7|WR47_ARATH Probable WRKY
transcription factor 47 (WRKY DNA-binding protein 47)
gi|25407124|pir||B85022 probable DNA-binding protein
[imported] - Arabidopsis thaliana
gi|7268215|emb|CAB77742.1| putative DNA-binding protein
[Arabidopsis thaliana]
gi|19172392|gb|AAL85881.1|AF480165_1 WRKY transcription
factor 47 [Arabidopsis thaliana]
Length = 489
Score = 55.8 bits (133), Expect = 4e-07
Identities = 40/83 (48%), Positives = 50/83 (60%)
Frame = +1
Query: 202 GTHNHPLPVSATAMDSSTSAALSML*SGSYTSQPTGHCAASMGSASTMFIGLNFSSYNQS 381
G HNHPLP SATAM ++TSAA +ML SGS +S ++ ++S+ F NF Y +
Sbjct: 292 GNHNHPLPPSATAMAATTSAAAAMLLSGSSSSNLHQTLSSPSATSSSSFYH-NF-PYTST 349
Query: 382 RATQAFLPTPTPHLFPTITLDLT 450
AT L P FPTITLDLT
Sbjct: 350 IAT---LSASAP--FPTITLDLT 367
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 512,874,958
Number of Sequences: 1393205
Number of extensions: 11283988
Number of successful extensions: 24246
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 23309
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24208
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)