Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002126A_C01 KMC002126A_c01
(592 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
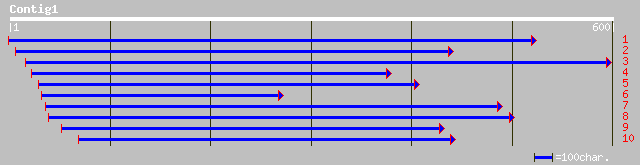
ref|NP_564748.1| expressed protein; protein id: At1g59600.1, sup... 115 3e-25
gb|AAF79773.1|AC009317_32 T30E16.16 [Arabidopsis thaliana] 98 7e-20
dbj|BAC20891.1| contains EST C27412(C51815)~similar to Arabidops... 92 6e-18
sp|Q46861|YGIQ_ECOLI Hypothetical protein ygiQ 33 2.7
ref|NP_708831.1| orf, conserved hypothetical protein [Shigella f... 33 2.7
>ref|NP_564748.1| expressed protein; protein id: At1g59600.1, supported by cDNA:
gi_6520226 [Arabidopsis thaliana]
gi|25455733|pir||T52430 hypothetical protein ZCW7
[imported] - Arabidopsis thaliana
gi|6520227|dbj|BAA87955.1| ZCW7 [Arabidopsis thaliana]
Length = 324
Score = 115 bits (289), Expect = 3e-25
Identities = 58/113 (51%), Positives = 77/113 (67%)
Frame = -1
Query: 592 DDFGKDANEDSGDLVSGLYCKIVVQFEKYIDFHDALRVLCSRSLQKQGSRLKADYEVSWD 413
DD GKDA+EDSGDL+SGL+CKIVVQFEKY DF +A++ C RS++K+G+RLKADYE++WD
Sbjct: 227 DDLGKDADEDSGDLISGLHCKIVVQFEKYNDFVNAMKAFCGRSMEKEGTRLKADYELTWD 286
Query: 412 KDGFFRYSRTQTQEKNNGVSKIATQQNRSEAPRRQSYNSRQSPDRVRPRRFQE 254
K GFFR SR ++ G R +S+ Q D +R +RF+E
Sbjct: 287 KIGFFRNSRKPLDNRDGGY-------------RNESFGCNQ--DDLRRKRFRE 324
>gb|AAF79773.1|AC009317_32 T30E16.16 [Arabidopsis thaliana]
Length = 383
Score = 98.2 bits (243), Expect = 7e-20
Identities = 54/136 (39%), Positives = 73/136 (52%), Gaps = 37/136 (27%)
Frame = -1
Query: 592 DDFGKDANEDSGDLVSGLYCKIVVQFEKYIDFHDALRVLCSRSLQK-------------- 455
DD GKDA+EDSGDL+SGL+CKIVVQFEKY DF +A++ C RS++K
Sbjct: 230 DDLGKDADEDSGDLISGLHCKIVVQFEKYNDFVNAMKAFCGRSMEKVFFMLQRPLSEMVL 289
Query: 454 -----------------------QGSRLKADYEVSWDKDGFFRYSRTQTQEKNNGVSKIA 344
+G+RLKADYE++WDK GFFR SR ++ G +
Sbjct: 290 LNASSEFGARLYAKLVSASVILWEGTRLKADYELTWDKIGFFRNSRKPLDNRDGGYRNES 349
Query: 343 TQQNRSEAPRRQSYNS 296
N+ + R++ NS
Sbjct: 350 FGCNQDDLRRKRFRNS 365
>dbj|BAC20891.1| contains EST C27412(C51815)~similar to Arabidopsis thaliana
chromosome 1,At1g59600~unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 340
Score = 91.7 bits (226), Expect = 6e-18
Identities = 46/113 (40%), Positives = 71/113 (62%)
Frame = -1
Query: 592 DDFGKDANEDSGDLVSGLYCKIVVQFEKYIDFHDALRVLCSRSLQKQGSRLKADYEVSWD 413
D++G + + +++SGL CK+ VQFE Y DF+ A++ LC RSL+K+GSRLK DYEV+WD
Sbjct: 236 DEWGAKQDGTNKEIISGLNCKVWVQFENYDDFNSAMQALCGRSLEKEGSRLKVDYEVTWD 295
Query: 412 KDGFFRYSRTQTQEKNNGVSKIATQQNRSEAPRRQSYNSRQSPDRVRPRRFQE 254
+GFFR ++ + N ++N S R++ Y SR D +RF++
Sbjct: 296 HEGFFRNAQYEPVRSN------LEERNSSAHGRKKHYTSRIESD--HRKRFRD 340
>sp|Q46861|YGIQ_ECOLI Hypothetical protein ygiQ
Length = 739
Score = 33.1 bits (74), Expect = 2.7
Identities = 26/92 (28%), Positives = 41/92 (44%), Gaps = 3/92 (3%)
Frame = -1
Query: 529 IVVQFEKYIDFHDALRVLCSRSLQKQGSRLKADYEVSW-DKDGFFRYSRTQTQEKNNGVS 353
I+V + Y+D +C R L+ QG R+ + W KD F R + GV+
Sbjct: 48 ILVTGDAYVDHPSFGMAICGRMLEAQGFRVGIIAQPDWSSKDDFMRLGKPNL---FFGVT 104
Query: 352 --KIATQQNRSEAPRRQSYNSRQSPDRVRPRR 263
+ + NR A RR ++ +PD V +R
Sbjct: 105 AGNMDSMINRYTADRRLRHDDAYTPDNVAGKR 136
>ref|NP_708831.1| orf, conserved hypothetical protein [Shigella flexneri 2a str. 301]
gi|24053480|gb|AAN44538.1|AE015318_4 orf, conserved
hypothetical protein [Shigella flexneri 2a str. 301]
Length = 739
Score = 33.1 bits (74), Expect = 2.7
Identities = 26/92 (28%), Positives = 41/92 (44%), Gaps = 3/92 (3%)
Frame = -1
Query: 529 IVVQFEKYIDFHDALRVLCSRSLQKQGSRLKADYEVSW-DKDGFFRYSRTQTQEKNNGVS 353
I+V + Y+D +C R L+ QG R+ + W KD F R + GV+
Sbjct: 48 ILVTGDAYVDHPSFGMAICGRMLEAQGFRVGIIAQPDWSSKDDFMRLGKPNL---FFGVT 104
Query: 352 --KIATQQNRSEAPRRQSYNSRQSPDRVRPRR 263
+ + NR A RR ++ +PD V +R
Sbjct: 105 AGNMDSMINRYTADRRLRHDDAYTPDNVAGKR 136
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 484,121,032
Number of Sequences: 1393205
Number of extensions: 10021344
Number of successful extensions: 24575
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 23955
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24572
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)