Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002124A_C01 KMC002124A_c01
(860 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
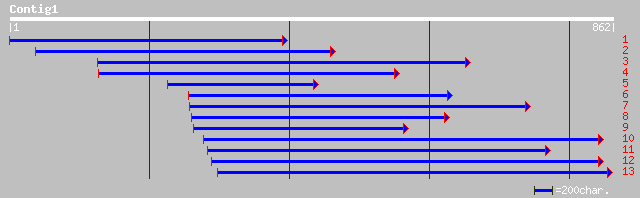
gb|AAM29182.1| Aux/IAA protein [Solanum tuberosum] 178 1e-43
emb|CAD10639.1| IAA9 protein [Nicotiana tabacum] 176 4e-43
gb|AAM12952.1| auxin-regulated protein [Zinnia elegans] 174 1e-42
pir||T05726 GH1 protein - soybean (fragment) gi|2388689|gb|AAB70... 172 6e-42
emb|CAC84706.1| aux/IAA protein [Populus tremula x Populus tremu... 169 4e-41
>gb|AAM29182.1| Aux/IAA protein [Solanum tuberosum]
Length = 349
Score = 178 bits (451), Expect = 1e-43
Identities = 86/112 (76%), Positives = 94/112 (83%)
Frame = -3
Query: 858 GAPYLRKVDLKTYKNYMEFSSALEKMFSCFTIGQCNSPGLPGKDGLSESSLRDLLHGSEY 679
GAPYLRKVDL+TY Y E SSALE MFSCFTIGQ S G PGKD LSES L+DLLHGSEY
Sbjct: 238 GAPYLRKVDLRTYSAYQELSSALETMFSCFTIGQYGSHGAPGKDMLSESKLKDLLHGSEY 297
Query: 678 VLTYEDKDGDWMLVGDVPWEMFTDSCRRLRIMKGSEAIGLELVERFECRSQH 523
VLTYEDKDGDWMLVGDVPWEMF D+C+RLRIMKGS+AIGL +CRS++
Sbjct: 298 VLTYEDKDGDWMLVGDVPWEMFIDTCKRLRIMKGSDAIGLAPRAMEKCRSRN 349
>emb|CAD10639.1| IAA9 protein [Nicotiana tabacum]
Length = 346
Score = 176 bits (446), Expect = 4e-43
Identities = 85/112 (75%), Positives = 94/112 (83%)
Frame = -3
Query: 858 GAPYLRKVDLKTYKNYMEFSSALEKMFSCFTIGQCNSPGLPGKDGLSESSLRDLLHGSEY 679
GAPYLRKVDLK Y Y E SSALEKMFSCFTIGQ S G PGK+ LSES L+DLLHGSEY
Sbjct: 235 GAPYLRKVDLKNYSAYQELSSALEKMFSCFTIGQYGSHGAPGKEMLSESQLKDLLHGSEY 294
Query: 678 VLTYEDKDGDWMLVGDVPWEMFTDSCRRLRIMKGSEAIGLELVERFECRSQH 523
VLTYEDKDGDWMLVGDVPWEMF ++C+RLRIMKGS+AIGL +CRS++
Sbjct: 295 VLTYEDKDGDWMLVGDVPWEMFIETCKRLRIMKGSDAIGLAPRGMEKCRSRN 346
>gb|AAM12952.1| auxin-regulated protein [Zinnia elegans]
Length = 351
Score = 174 bits (442), Expect = 1e-42
Identities = 79/100 (79%), Positives = 89/100 (89%)
Frame = -3
Query: 858 GAPYLRKVDLKTYKNYMEFSSALEKMFSCFTIGQCNSPGLPGKDGLSESSLRDLLHGSEY 679
GAPYLRKVDL++Y Y + SSA+EKMFSCFTIGQC S G PG++ LSES LRDLLHGSEY
Sbjct: 240 GAPYLRKVDLRSYSTYQQLSSAIEKMFSCFTIGQCGSQGAPGRESLSESKLRDLLHGSEY 299
Query: 678 VLTYEDKDGDWMLVGDVPWEMFTDSCRRLRIMKGSEAIGL 559
VLTYEDKDGDWMLVGDVPW+MF SC+RL+IMKGS+AIGL
Sbjct: 300 VLTYEDKDGDWMLVGDVPWDMFIGSCKRLKIMKGSDAIGL 339
>pir||T05726 GH1 protein - soybean (fragment) gi|2388689|gb|AAB70005.1| GH1
protein [Glycine max]
Length = 339
Score = 172 bits (436), Expect = 6e-42
Identities = 84/113 (74%), Positives = 96/113 (84%), Gaps = 3/113 (2%)
Frame = -3
Query: 858 GAPYLRKVDLKTYKNYMEFSSALEKMF-SCFTIGQCNSPGLPGKDGLSESSLRDLLHGSE 682
GAPYLRKVDL++Y Y E SSALEKMF SCFT+GQC S G PG++ LSES LRDLLHGSE
Sbjct: 227 GAPYLRKVDLRSYTTYQELSSALEKMFLSCFTLGQCGSHGAPGREMLSESKLRDLLHGSE 286
Query: 681 YVLTYEDKDGDWMLVGDVPWEMFTDSCRRLRIMKGSEAIGL--ELVERFECRS 529
YVLTYEDKDGDWMLVGDVPWEMF D+C+RL+IMKGS+AIGL +E+ + RS
Sbjct: 287 YVLTYEDKDGDWMLVGDVPWEMFIDTCKRLKIMKGSDAIGLAPRAMEKSKSRS 339
>emb|CAC84706.1| aux/IAA protein [Populus tremula x Populus tremuloides]
Length = 365
Score = 169 bits (429), Expect = 4e-41
Identities = 80/112 (71%), Positives = 93/112 (82%)
Frame = -3
Query: 858 GAPYLRKVDLKTYKNYMEFSSALEKMFSCFTIGQCNSPGLPGKDGLSESSLRDLLHGSEY 679
GAPYLRKVDL+ Y Y E SSALEKMFSCFTIGQ S G PG++ LSES L+DLLHGSEY
Sbjct: 254 GAPYLRKVDLRNYSAYQELSSALEKMFSCFTIGQYGSHGAPGREMLSESKLKDLLHGSEY 313
Query: 678 VLTYEDKDGDWMLVGDVPWEMFTDSCRRLRIMKGSEAIGLELVERFECRSQH 523
VLTYEDKDGDWMLVGDVPWEMF ++C+RLRIMK S+AIGL +C++++
Sbjct: 314 VLTYEDKDGDWMLVGDVPWEMFIETCKRLRIMKSSDAIGLAPRAMEKCKNRN 365
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 752,839,257
Number of Sequences: 1393205
Number of extensions: 17060355
Number of successful extensions: 45095
Number of sequences better than 10.0: 199
Number of HSP's better than 10.0 without gapping: 41092
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44884
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 45709790868
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)