Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002123A_C04 KMC002123A_c04
(577 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
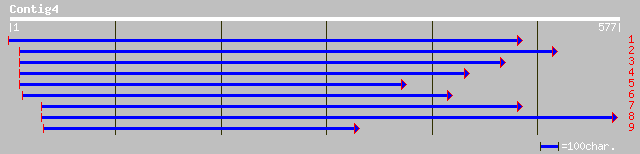
gb|AAM66053.1| copia-like retroelement pol polyprotein [Arabidop... 116 2e-25
ref|NP_194645.1| putative protein; protein id: At4g29160.1, supp... 116 2e-25
ref|NP_179573.1| copia-like retroelement pol polyprotein; protei... 98 7e-20
gb|AAH45919.1| Unknown (protein for MGC:56112) [Danio rerio] 63 3e-09
gb|AAH44191.1| Similar to RIKEN cDNA 2010012F05 gene [Danio rerio] 56 4e-07
>gb|AAM66053.1| copia-like retroelement pol polyprotein [Arabidopsis thaliana]
Length = 218
Score = 116 bits (290), Expect = 2e-25
Identities = 63/81 (77%), Positives = 67/81 (81%), Gaps = 1/81 (1%)
Frame = -1
Query: 577 IQEALSAPIGAAADFDEDELEAELEELEGAELEEQLLQPATTA-PAASVHVPAGRQPTRP 401
IQEAL+ P+GAAADFDEDEL AEL+ELE ELE QLLQPATTA P SV VPAGRQP RP
Sbjct: 138 IQEALATPMGAAADFDEDELAAELDELESEELESQLLQPATTAPPLPSVPVPAGRQPARP 197
Query: 400 VSAKTTPEEDELAALQAEMAL 338
V K T EE+ELAALQAEMAL
Sbjct: 198 VPQKRTAEEEELAALQAEMAL 218
>ref|NP_194645.1| putative protein; protein id: At4g29160.1, supported by cDNA:
7471., supported by cDNA: gi_14596130, supported by
cDNA: gi_18377477 [Arabidopsis thaliana]
gi|7485746|pir||T08971 hypothetical protein F19B15.190 -
Arabidopsis thaliana gi|4972062|emb|CAB43930.1| putative
protein [Arabidopsis thaliana]
gi|7269814|emb|CAB79674.1| putative protein [Arabidopsis
thaliana] gi|14596131|gb|AAK68793.1| putative protein
[Arabidopsis thaliana] gi|18377478|gb|AAL66905.1|
putative protein [Arabidopsis thaliana]
Length = 219
Score = 116 bits (290), Expect = 2e-25
Identities = 63/81 (77%), Positives = 67/81 (81%), Gaps = 1/81 (1%)
Frame = -1
Query: 577 IQEALSAPIGAAADFDEDELEAELEELEGAELEEQLLQPATTA-PAASVHVPAGRQPTRP 401
IQEAL+ P+GAAADFDEDEL AEL+ELE ELE QLLQPATTA P SV VPAGRQP RP
Sbjct: 139 IQEALATPMGAAADFDEDELAAELDELESEELESQLLQPATTAPPLPSVPVPAGRQPARP 198
Query: 400 VSAKTTPEEDELAALQAEMAL 338
V K T EE+ELAALQAEMAL
Sbjct: 199 VPQKRTAEEEELAALQAEMAL 219
>ref|NP_179573.1| copia-like retroelement pol polyprotein; protein id: At2g19830.1,
supported by cDNA: gi_15028176, supported by cDNA:
gi_19310842 [Arabidopsis thaliana]
gi|25370522|pir||F84581 copia-like retroelement pol
polyprotein [imported] - Arabidopsis thaliana
gi|3687235|gb|AAC62133.1| copia-like retroelement pol
polyprotein [Arabidopsis thaliana]
gi|15028177|gb|AAK76585.1| putative copia retroelement
pol polyprotein [Arabidopsis thaliana]
gi|19310843|gb|AAL85152.1| putative copia retroelement
pol polyprotein [Arabidopsis thaliana]
Length = 213
Score = 98.2 bits (243), Expect = 7e-20
Identities = 57/81 (70%), Positives = 61/81 (74%), Gaps = 1/81 (1%)
Frame = -1
Query: 577 IQEALSAPIGAAADFDEDELEAELEELEGAELEEQLLQPATTAPAASVHVPAGRQPTR-P 401
IQEALSAP GA DFDEDELEAEL+ELEGAELEEQLLQP +HVP G +P R P
Sbjct: 140 IQEALSAPFGAN-DFDEDELEAELDELEGAELEEQLLQP------VPIHVPQGNKPARAP 192
Query: 400 VSAKTTPEEDELAALQAEMAL 338
+ T EEDELAALQAEMAL
Sbjct: 193 AQKQPTAEEDELAALQAEMAL 213
>gb|AAH45919.1| Unknown (protein for MGC:56112) [Danio rerio]
Length = 220
Score = 62.8 bits (151), Expect = 3e-09
Identities = 33/76 (43%), Positives = 47/76 (61%)
Frame = -1
Query: 577 IQEALSAPIGAAADFDEDELEAELEELEGAELEEQLLQPATTAPAASVHVPAGRQPTRPV 398
I +A+S P+G +FDEDEL AELEELE EL++ LL+ P +VP+ P+RP
Sbjct: 143 ISDAISKPVGFGEEFDEDELLAELEELEQEELDKNLLEIGDNVPLP--NVPSTSLPSRPA 200
Query: 397 SAKTTPEEDELAALQA 350
K +ED++ L+A
Sbjct: 201 KKKEEEDEDDMKDLEA 216
>gb|AAH44191.1| Similar to RIKEN cDNA 2010012F05 gene [Danio rerio]
Length = 224
Score = 55.8 bits (133), Expect = 4e-07
Identities = 32/71 (45%), Positives = 43/71 (60%), Gaps = 1/71 (1%)
Frame = -1
Query: 577 IQEALSAPIGAAADFDEDELEAELEELEGAELEEQLLQPATTAPAASVHVPAGRQPTR-P 401
I +A+S P+G +FDEDEL AELEELE EL++ LLQ + +VP+ P +
Sbjct: 143 ISDAISRPVGFGEEFDEDELMAELEELEQEELDKDLLQISGPEDVPLPNVPSNPLPKKTA 202
Query: 400 VSAKTTPEEDE 368
V+ K EEDE
Sbjct: 203 VAQKKRQEEDE 213
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 497,206,906
Number of Sequences: 1393205
Number of extensions: 11086596
Number of successful extensions: 81710
Number of sequences better than 10.0: 681
Number of HSP's better than 10.0 without gapping: 46444
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 64094
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)