Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
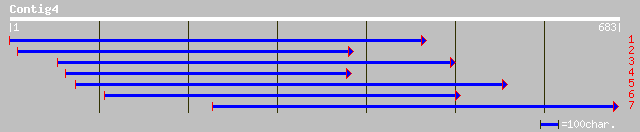
Query= KMC002089A_C04 KMC002089A_c04
(683 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565711.1| expressed protein; protein id: At2g31040.1, sup... 237 1e-61
gb|AAM64960.1| unknown [Arabidopsis thaliana] 237 1e-61
dbj|BAB84384.1| P0007F06.23 [Oryza sativa (japonica cultivar-gro... 192 3e-48
ref|NP_484055.1| ATP synthase subunit 1 [Nostoc sp. PCC 7120] gi... 55 9e-07
ref|ZP_00074541.1| hypothetical protein [Trichodesmium erythraeu... 55 1e-06
>ref|NP_565711.1| expressed protein; protein id: At2g31040.1, supported by cDNA:
35095., supported by cDNA: gi_15215860, supported by
cDNA: gi_19699267 [Arabidopsis thaliana]
gi|25370752|pir||G84715 hypothetical protein At2g31040
[imported] - Arabidopsis thaliana
gi|3746067|gb|AAC63842.1| expressed protein [Arabidopsis
thaliana] gi|15215861|gb|AAK91474.1| At2g31040/T16B12.15
[Arabidopsis thaliana] gi|19699268|gb|AAL91000.1|
At2g31040/T16B12.15 [Arabidopsis thaliana]
gi|20197222|gb|AAM14978.1| expressed protein
[Arabidopsis thaliana]
Length = 350
Score = 237 bits (604), Expect = 1e-61
Identities = 118/155 (76%), Positives = 137/155 (88%)
Frame = -1
Query: 683 DVMFRELRRPRENPEVLAAQAEEQYDKLKNKMQILTLGIGGVGLVSSYVSYSPEIAASFG 504
DVMFRELRRPR +PEV AA+ EQY KLKNK+Q+LTLGIGGVGLVS+Y+SY+PEIA SFG
Sbjct: 185 DVMFRELRRPRGDPEVQAAKDREQYFKLKNKIQVLTLGIGGVGLVSAYISYTPEIALSFG 244
Query: 503 AGLLGSLAYIRMLGSSVDALRTNGGKGFVKGAIGQPRLLVPVILVMVYNRWNAILVPEFG 324
AGLLGSLAY+RMLG+SVDA+ +G +G KGA QPRLLVPV+LVM++NRWNAILVPE+G
Sbjct: 245 AGLLGSLAYMRMLGNSVDAM-ADGARGVAKGAANQPRLLVPVVLVMIFNRWNAILVPEYG 303
Query: 323 AMHLELIPMLVGFFTYQIGTFFPAIEEAITIAVKK 219
MHLELIPMLVGFFTY+I TFF AIEEAI+I +K
Sbjct: 304 FMHLELIPMLVGFFTYKIATFFQAIEEAISITTQK 338
>gb|AAM64960.1| unknown [Arabidopsis thaliana]
Length = 350
Score = 237 bits (604), Expect = 1e-61
Identities = 118/155 (76%), Positives = 137/155 (88%)
Frame = -1
Query: 683 DVMFRELRRPRENPEVLAAQAEEQYDKLKNKMQILTLGIGGVGLVSSYVSYSPEIAASFG 504
DVMFRELRRPR +PEV AA+ EQY KLKNK+Q+LTLGIGGVGLVS+Y+SY+PEIA SFG
Sbjct: 185 DVMFRELRRPRGDPEVQAAKDREQYFKLKNKIQVLTLGIGGVGLVSAYISYTPEIALSFG 244
Query: 503 AGLLGSLAYIRMLGSSVDALRTNGGKGFVKGAIGQPRLLVPVILVMVYNRWNAILVPEFG 324
AGLLGSLAY+RMLG+SVDA+ +G +G KGA QPRLLVPV+LVM++NRWNAILVPE+G
Sbjct: 245 AGLLGSLAYMRMLGNSVDAM-ADGARGVAKGAANQPRLLVPVVLVMIFNRWNAILVPEYG 303
Query: 323 AMHLELIPMLVGFFTYQIGTFFPAIEEAITIAVKK 219
MHLELIPMLVGFFTY+I TFF AIEEAI+I +K
Sbjct: 304 FMHLELIPMLVGFFTYKIATFFQAIEEAISITTQK 338
>dbj|BAB84384.1| P0007F06.23 [Oryza sativa (japonica cultivar-group)]
gi|21644622|dbj|BAC01181.1| P0485G01.15 [Oryza sativa
(japonica cultivar-group)]
Length = 311
Score = 192 bits (489), Expect = 3e-48
Identities = 101/152 (66%), Positives = 124/152 (81%), Gaps = 3/152 (1%)
Frame = -1
Query: 683 DVMFREL--RRPRENPEVLAAQAEEQYDKLKNKMQILTLGIGGVGLVSSYVSYSPEIAAS 510
DV+ RE R + +PEVLAA++ EQY +LK ++Q+ TLGIGG+GLVS+Y SYSPEIAAS
Sbjct: 154 DVILRESKSRGQQGDPEVLAAKSREQYLELKQRLQLFTLGIGGIGLVSAYFSYSPEIAAS 213
Query: 509 FGAGLLGSLAYIRMLGSSVDALRTNGGKG-FVKGAIGQPRLLVPVILVMVYNRWNAILVP 333
FGAGL+GS+ Y+RMLG+SVD+L GG G VK A QPRLL+PV LVM+YNRWN ILVP
Sbjct: 214 FGAGLIGSVLYLRMLGTSVDSLA--GGTGETVKSAAAQPRLLIPVALVMMYNRWNEILVP 271
Query: 332 EFGAMHLELIPMLVGFFTYQIGTFFPAIEEAI 237
++G MHLELIPMLVGFFTY+I TF AI+E+I
Sbjct: 272 DYGFMHLELIPMLVGFFTYKIATFAQAIQESI 303
>ref|NP_484055.1| ATP synthase subunit 1 [Nostoc sp. PCC 7120]
gi|20141204|sp|P12403|ATPZ_ANASP ATP synthase protein I
gi|25296872|pir||AC1808 ATP synthase chain 1 [imported]
- Nostoc sp. (strain PCC 7120)
gi|17134989|dbj|BAB77535.1| ATP synthase subunit 1
[Nostoc sp. PCC 7120]
Length = 122
Score = 55.1 bits (131), Expect = 9e-07
Identities = 31/115 (26%), Positives = 63/115 (53%)
Frame = -1
Query: 617 EQYDKLKNKMQILTLGIGGVGLVSSYVSYSPEIAASFGAGLLGSLAYIRMLGSSVDALRT 438
+++ +L ++ ++TL + GV +S ++ YS IA ++ G + Y+RML V+ L
Sbjct: 2 QEFYQLYQELVLITLVLTGVVFISVWIFYSLNIALNYLLGACTGVVYLRMLAKDVERL-- 59
Query: 437 NGGKGFVKGAIGQPRLLVPVILVMVYNRWNAILVPEFGAMHLELIPMLVGFFTYQ 273
G K ++ + RL + + L+++ +RWN L+++P+ +GF TY+
Sbjct: 60 ----GREKQSLSKTRLALLMALILLASRWN----------QLQIMPIFLGFLTYK 100
>ref|ZP_00074541.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 146
Score = 54.7 bits (130), Expect = 1e-06
Identities = 33/115 (28%), Positives = 63/115 (54%)
Frame = -1
Query: 617 EQYDKLKNKMQILTLGIGGVGLVSSYVSYSPEIAASFGAGLLGSLAYIRMLGSSVDALRT 438
++Y KL+ ++ ++TL I G+ + +V YS IA ++ G S+ Y+RML V+ +
Sbjct: 32 KEYYKLQEELYVITLTITGIIFIFVWVFYSLNIALNYLIGATTSVVYLRMLAKDVERI-- 89
Query: 437 NGGKGFVKGAIGQPRLLVPVILVMVYNRWNAILVPEFGAMHLELIPMLVGFFTYQ 273
G KG++ + RL + V L+++ + N L+++P+ +GF TY+
Sbjct: 90 ----GREKGSLSKTRLAILVGLIILAAQLN----------ELKILPIFLGFLTYK 130
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 602,393,495
Number of Sequences: 1393205
Number of extensions: 14125347
Number of successful extensions: 38410
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 37030
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38374
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30552968016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)