Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002085A_C01 KMC002085A_c01
(533 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
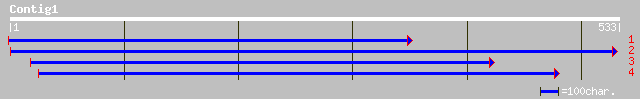
ref|NP_565615.1| G-protein beta family; protein id: At2g26060.1,... 97 7e-23
ref|NP_195025.2| G-protein beta family; protein id: At4g32990.1 ... 104 8e-22
pir||T02617 hypothetical protein At2g26060 [imported] - Arabidop... 68 2e-14
pir||T05307 hypothetical protein F26P21.110 - Arabidopsis thalia... 75 4e-13
ref|NP_610996.1| CG12797-PA [Drosophila melanogaster] gi|7303130... 59 3e-10
>ref|NP_565615.1| G-protein beta family; protein id: At2g26060.1, supported by cDNA:
142426., supported by cDNA: gi_20260509 [Arabidopsis
thaliana] gi|20197268|gb|AAC31230.2| expressed protein
[Arabidopsis thaliana] gi|20260510|gb|AAM13153.1|
unknown protein [Arabidopsis thaliana]
gi|21553416|gb|AAM62509.1| unknown [Arabidopsis
thaliana] gi|28059424|gb|AAO30057.1| unknown protein
[Arabidopsis thaliana]
Length = 352
Score = 96.7 bits (239), Expect(2) = 7e-23
Identities = 43/65 (66%), Positives = 49/65 (75%)
Frame = -3
Query: 531 GYHDRTIFSIHWSREGIFASGAADDTIQLFGDDNESQVGGPLYTLLLKKEKAHDMDINSV 352
GYHDRTI+S HWSR+ I ASGA D+ I+LF D V GP Y LLLKK KAH+ D+NSV
Sbjct: 264 GYHDRTIYSAHWSRDDIIASGAGDNAIRLFVDSKHDSVDGPSYNLLLKKNKAHENDVNSV 323
Query: 351 QWSPG 337
QWSPG
Sbjct: 324 QWSPG 328
Score = 32.0 bits (71), Expect(2) = 7e-23
Identities = 12/19 (63%), Positives = 17/19 (89%)
Frame = -1
Query: 299 LLASASDDGTIKVWELVSQ 243
LLASASDDG +K+W+L ++
Sbjct: 333 LLASASDDGMVKIWQLATK 351
>ref|NP_195025.2| G-protein beta family; protein id: At4g32990.1 [Arabidopsis
thaliana]
Length = 318
Score = 104 bits (259), Expect = 8e-22
Identities = 44/64 (68%), Positives = 54/64 (83%)
Frame = -3
Query: 531 GYHDRTIFSIHWSREGIFASGAADDTIQLFGDDNESQVGGPLYTLLLKKEKAHDMDINSV 352
G+HDRTI+S+HWSR+G+ ASGA DDTIQLF D + V GP Y LL+KKEKAH+MD+NSV
Sbjct: 240 GFHDRTIYSVHWSRDGVIASGAGDDTIQLFVDSDSDSVDGPSYKLLVKKEKAHEMDVNSV 299
Query: 351 QWSP 340
QW+P
Sbjct: 300 QWAP 303
>pir||T02617 hypothetical protein At2g26060 [imported] - Arabidopsis thaliana
Length = 323
Score = 67.8 bits (164), Expect(2) = 2e-14
Identities = 31/51 (60%), Positives = 36/51 (69%)
Frame = -3
Query: 489 EGIFASGAADDTIQLFGDDNESQVGGPLYTLLLKKEKAHDMDINSVQWSPG 337
+ I ASGA D+ I+LF D V GP Y LLLKK KAH+ D+NSVQWSPG
Sbjct: 249 DDIIASGAGDNAIRLFVDSKHDSVDGPSYNLLLKKNKAHENDVNSVQWSPG 299
Score = 32.0 bits (71), Expect(2) = 2e-14
Identities = 12/19 (63%), Positives = 17/19 (89%)
Frame = -1
Query: 299 LLASASDDGTIKVWELVSQ 243
LLASASDDG +K+W+L ++
Sbjct: 304 LLASASDDGMVKIWQLATK 322
>pir||T05307 hypothetical protein F26P21.110 - Arabidopsis thaliana
gi|3688180|emb|CAA21208.1| putative protein [Arabidopsis
thaliana] gi|7270246|emb|CAB80016.1| putative protein
[Arabidopsis thaliana]
Length = 243
Score = 75.5 bits (184), Expect = 4e-13
Identities = 33/50 (66%), Positives = 40/50 (80%)
Frame = -3
Query: 489 EGIFASGAADDTIQLFGDDNESQVGGPLYTLLLKKEKAHDMDINSVQWSP 340
+G+ ASGA DDTIQLF D + V GP Y LL+KKEKAH+MD+NSVQW+P
Sbjct: 179 DGVIASGAGDDTIQLFVDSDSDSVDGPSYKLLVKKEKAHEMDVNSVQWAP 228
>ref|NP_610996.1| CG12797-PA [Drosophila melanogaster] gi|7303130|gb|AAF58195.1|
CG12797-PA [Drosophila melanogaster]
gi|16769434|gb|AAL28936.1| LD31217p [Drosophila
melanogaster]
Length = 335
Score = 58.5 bits (140), Expect(2) = 3e-10
Identities = 24/65 (36%), Positives = 40/65 (60%), Gaps = 1/65 (1%)
Frame = -3
Query: 531 GYHDRTIFSIHWSR-EGIFASGAADDTIQLFGDDNESQVGGPLYTLLLKKEKAHDMDINS 355
G H R I+ + W + G+ A+ DD I++F + ++S+ P + + +E AHD D+NS
Sbjct: 249 GQHSRAIYDVSWCKLTGLIATACGDDGIRIFKESSDSKPDEPTFEQITAEEGAHDQDVNS 308
Query: 354 VQWSP 340
VQW+P
Sbjct: 309 VQWNP 313
Score = 27.3 bits (59), Expect(2) = 3e-10
Identities = 10/15 (66%), Positives = 13/15 (86%)
Frame = -1
Query: 296 LASASDDGTIKVWEL 252
L S SDDGTIK+W++
Sbjct: 319 LISCSDDGTIKIWKV 333
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 440,559,070
Number of Sequences: 1393205
Number of extensions: 9383867
Number of successful extensions: 31807
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 28697
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31787
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17885181664
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)